Engineering of ultraID, a compact and hyperactive enzyme for proximity-dependent biotinylation in living cells | Communications Biology

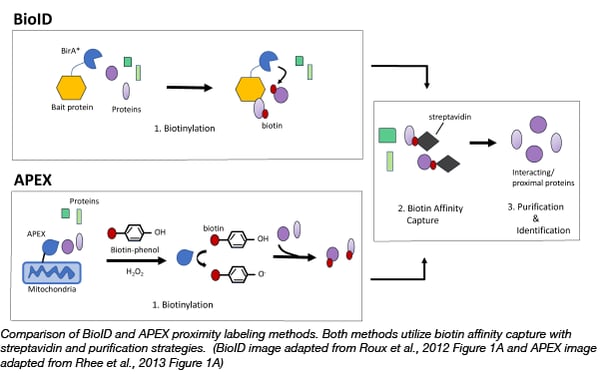
Cells | Free Full-Text | Comparative Application of BioID and TurboID for Protein-Proximity Biotinylation

Probing mammalian centrosome structure using BioID proximity-dependent biotinylation - ScienceDirect

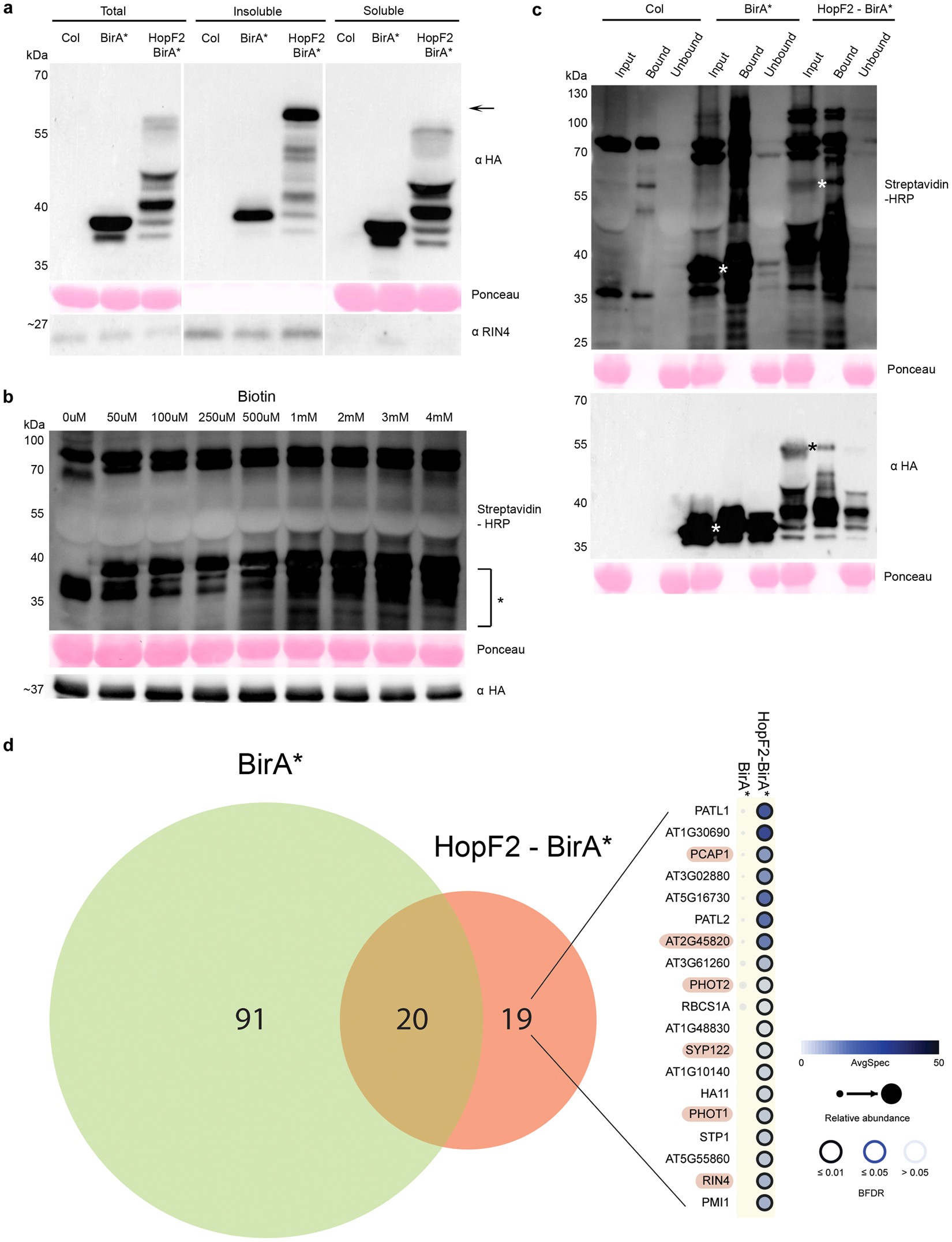
In planta proximity-dependent biotin identification (BioID) identifies a TMV replication co-chaperone NbSGT1 in the vicinity of 126 kDa replicase - ScienceDirect

Split-Bioid — Proteomic Analysis Of Context-Specific Protein Complexes In Their Native Cellular Environment - Video
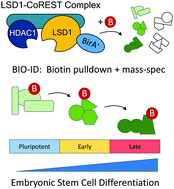
Proximity-dependent biotin identification (BioID) reveals a dynamic LSD1–CoREST interactome during embryonic stem cell differentiation - Molecular Omics (RSC Publishing)
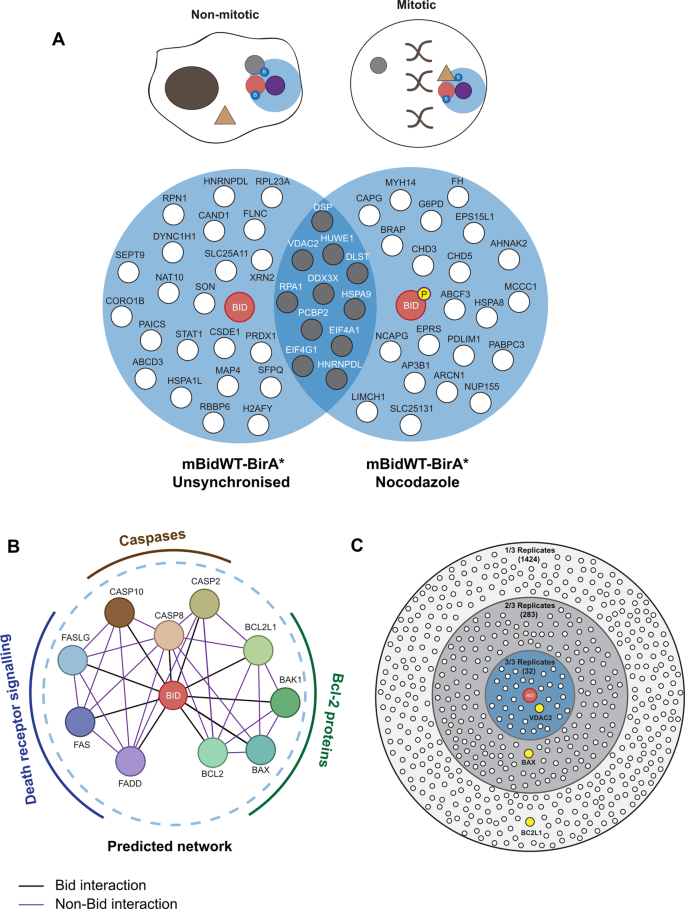
BioID-based proteomic analysis of the Bid interactome identifies novel proteins involved in cell-cycle-dependent apoptotic priming | Cell Death & Disease

Model for application of BioID method. (a) Expression of a promiscuous... | Download Scientific Diagram
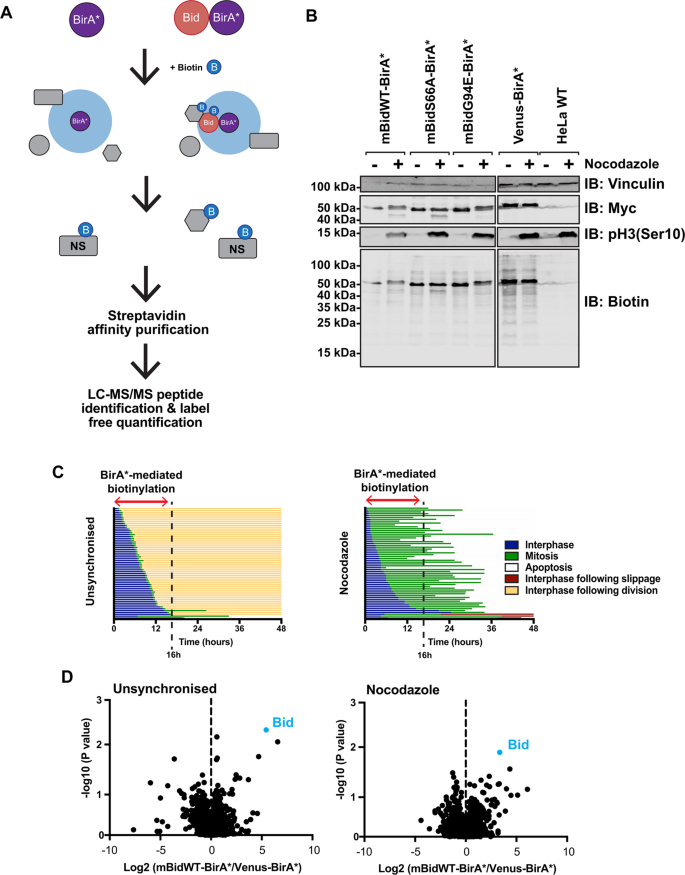
BioID-based proteomic analysis of the Bid interactome identifies novel proteins involved in cell-cycle-dependent apoptotic priming | Cell Death & Disease

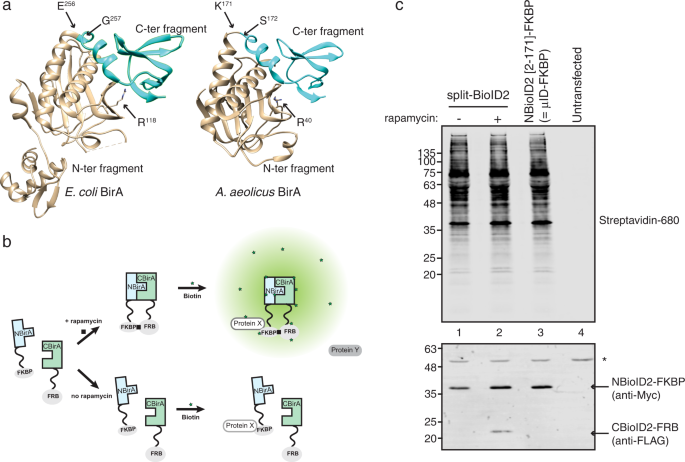
Identification of interacting proteins using proximity-dependent biotinylation with BioID2 in Trypanosoma brucei
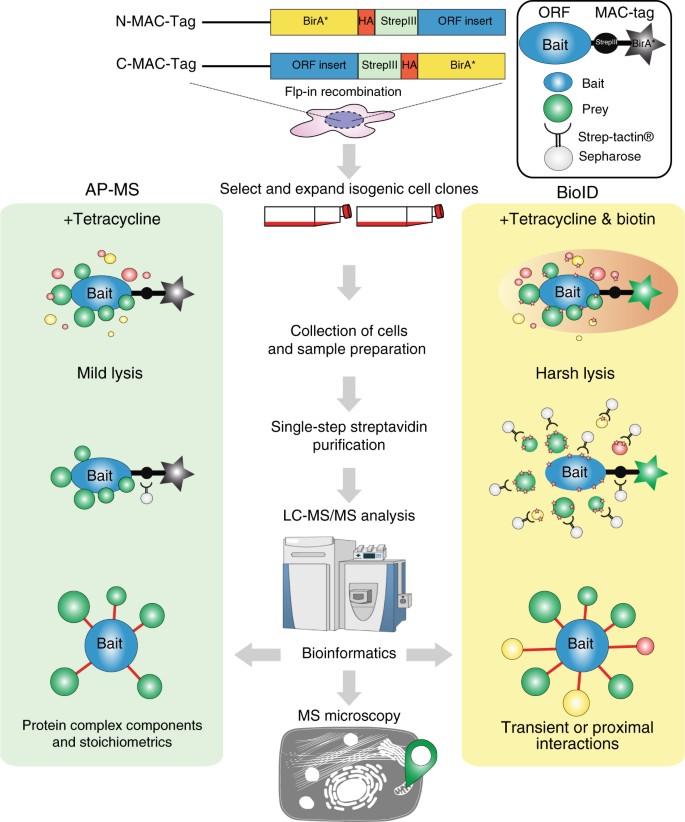
An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations | Nature Communications

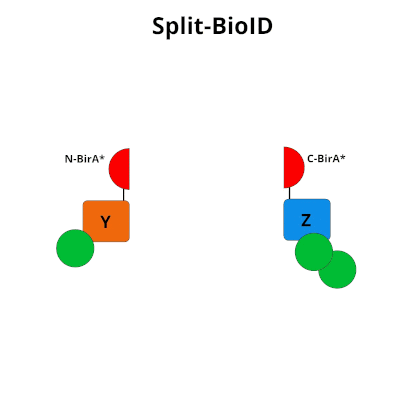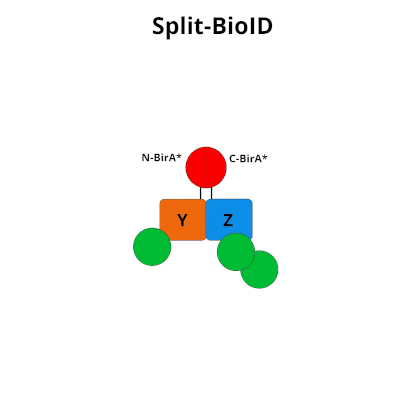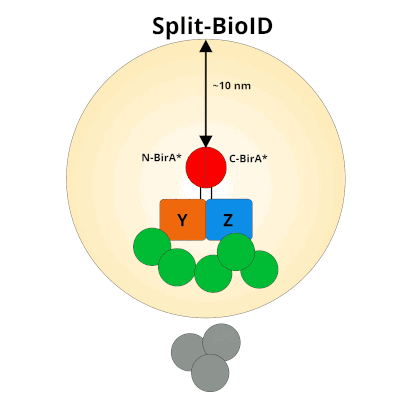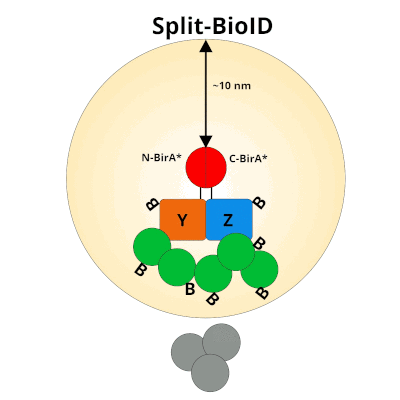
Split‐BioID: a proximity biotinylation assay for dimerization‐dependent protein interactions - De Munter - 2017 - FEBS Letters - Wiley Online Library