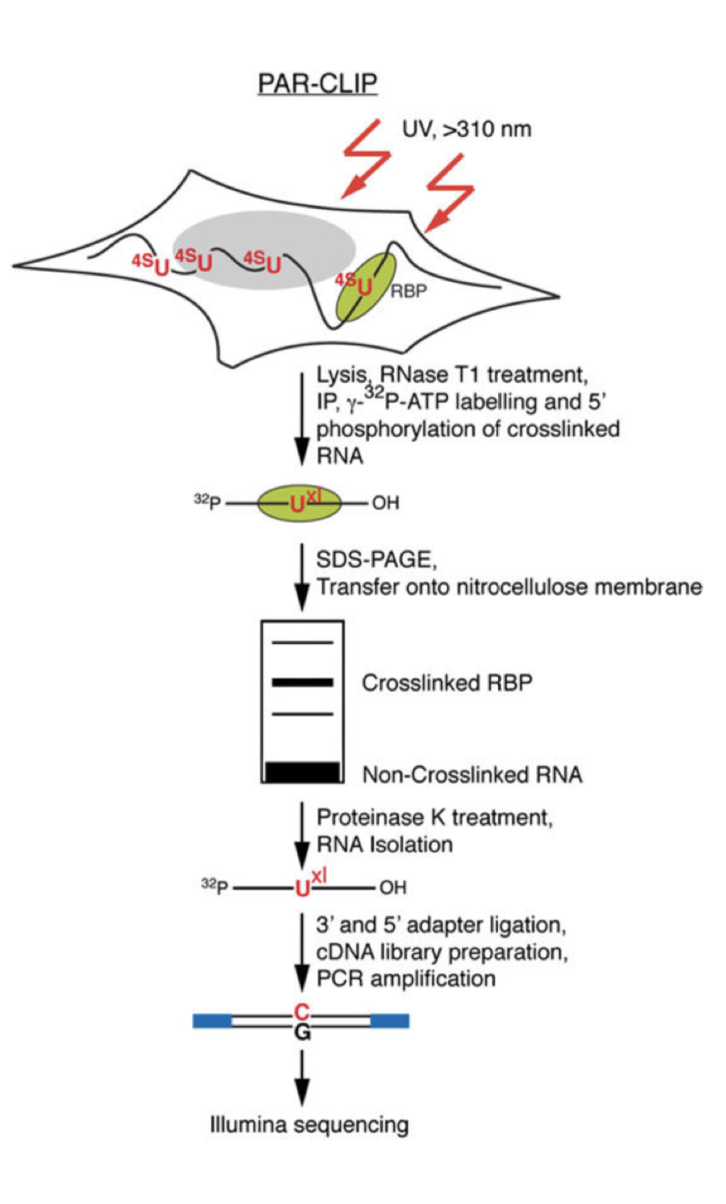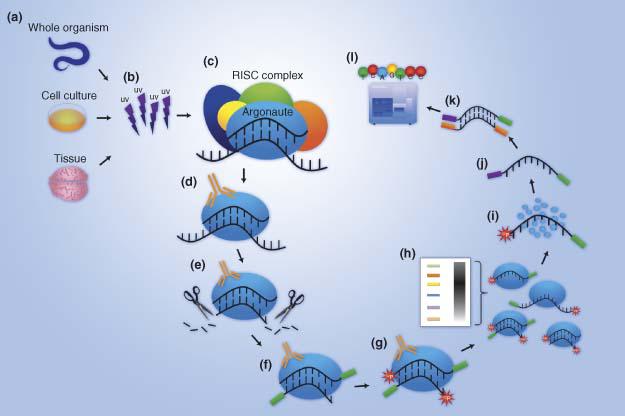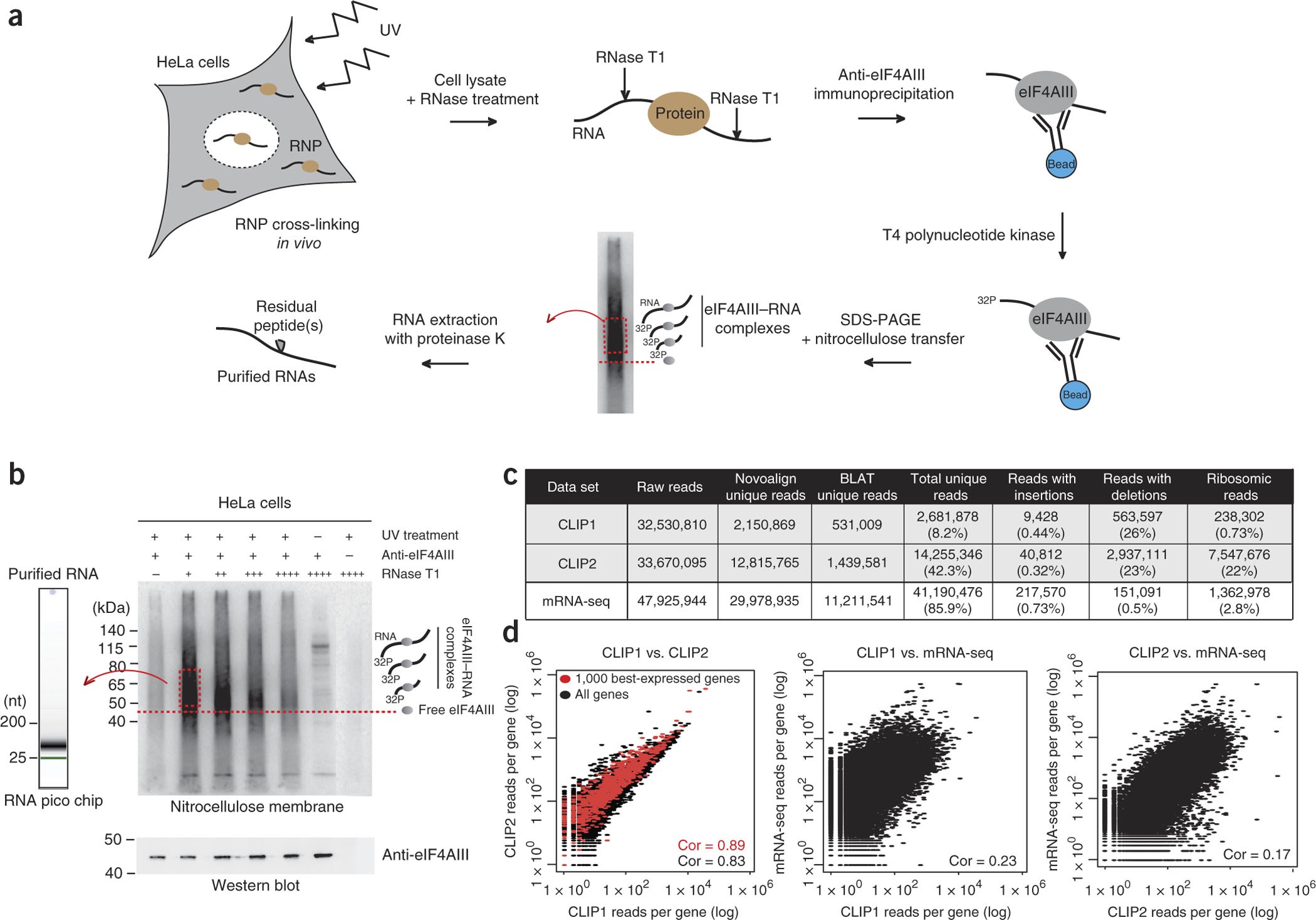
CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex | Nature Structural & Molecular Biology
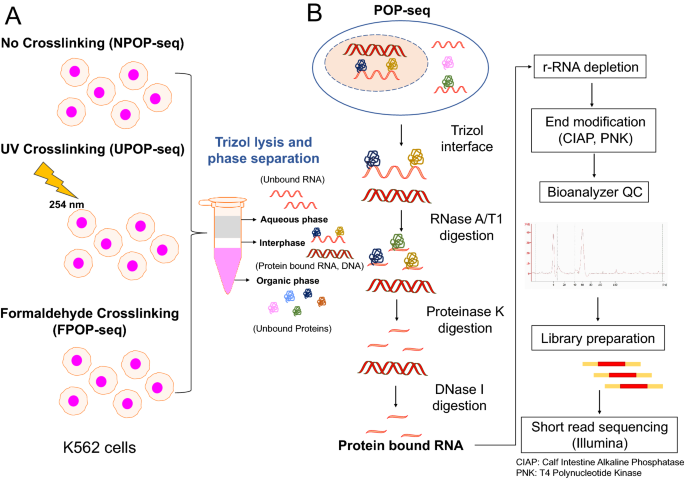
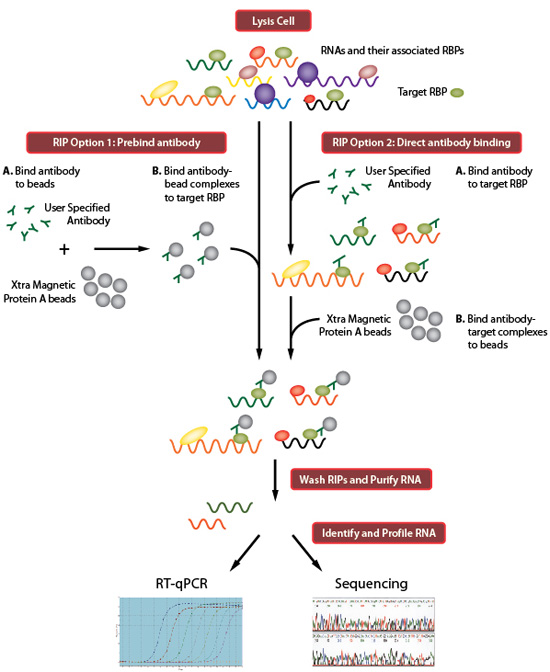
Transcriptome-wide high-throughput mapping of protein–RNA occupancy profiles using POP-seq | Scientific Reports
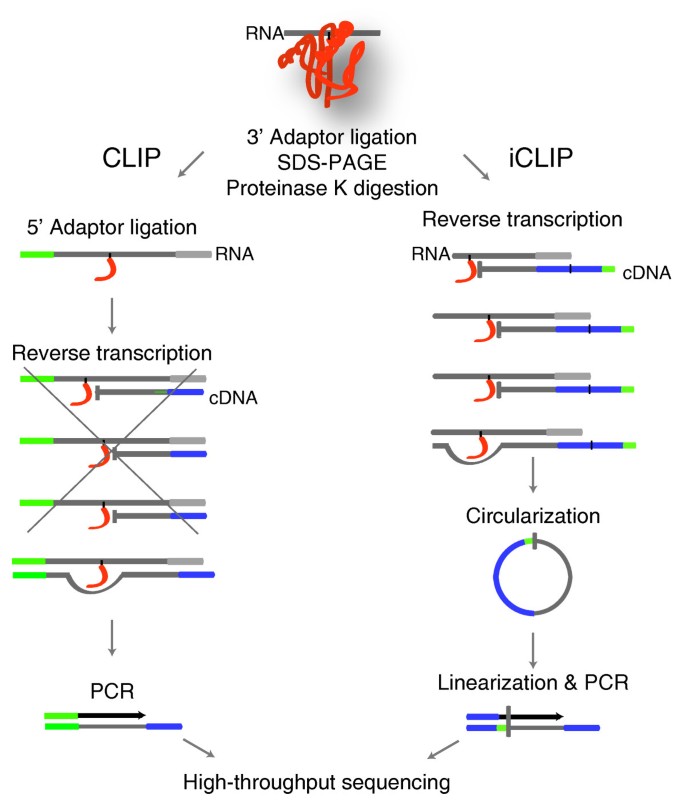
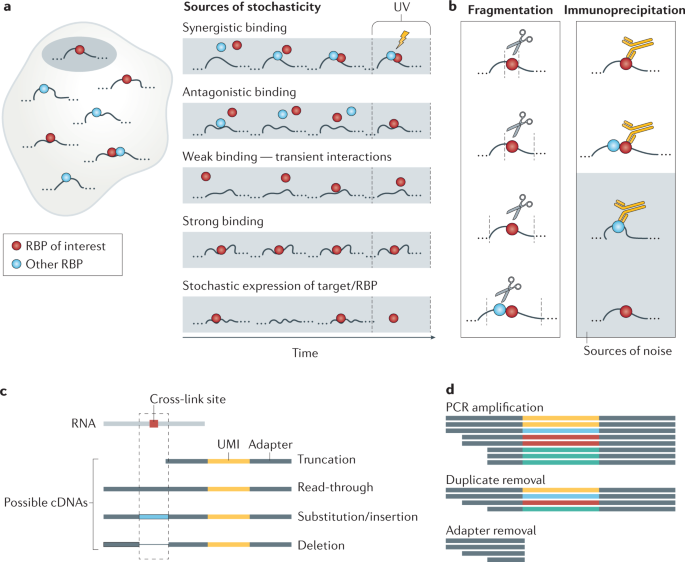
Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions | Genome Biology | Full Text
Learning to Predict miRNA-mRNA Interactions from AGO CLIP Sequencing and CLASH Data | PLOS Computational Biology

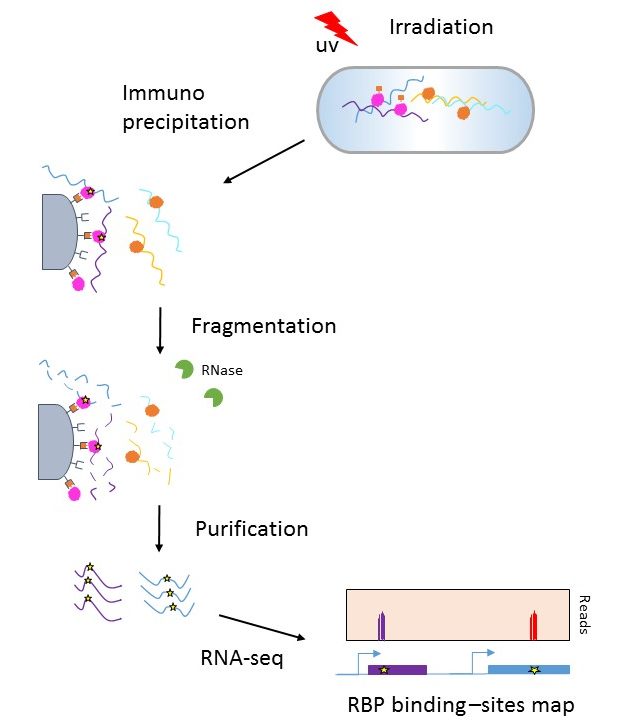
Flow diagram of CLIP-seq. Cells are fed with the ribonucleoside analog... | Download Scientific Diagram

Main steps of the bioinformatics workflow to analyze CLIP-seq data with... | Download Scientific Diagram

Computational approaches for the analysis of RNA–protein interactions: A primer for biologists - ScienceDirect
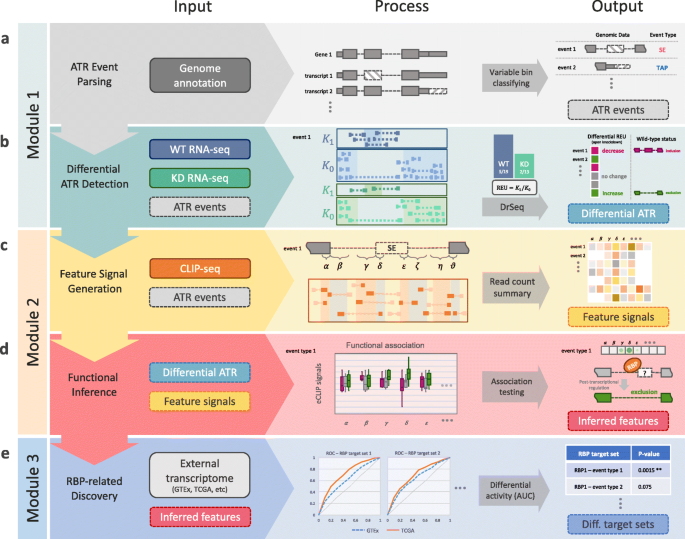
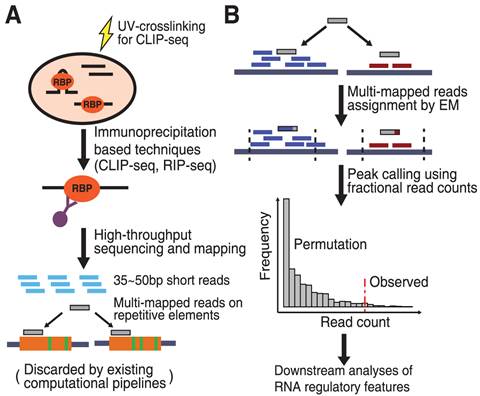
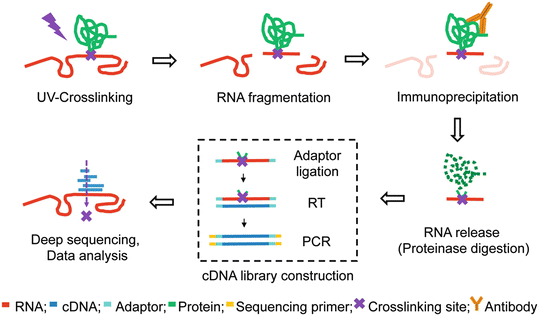
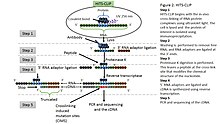
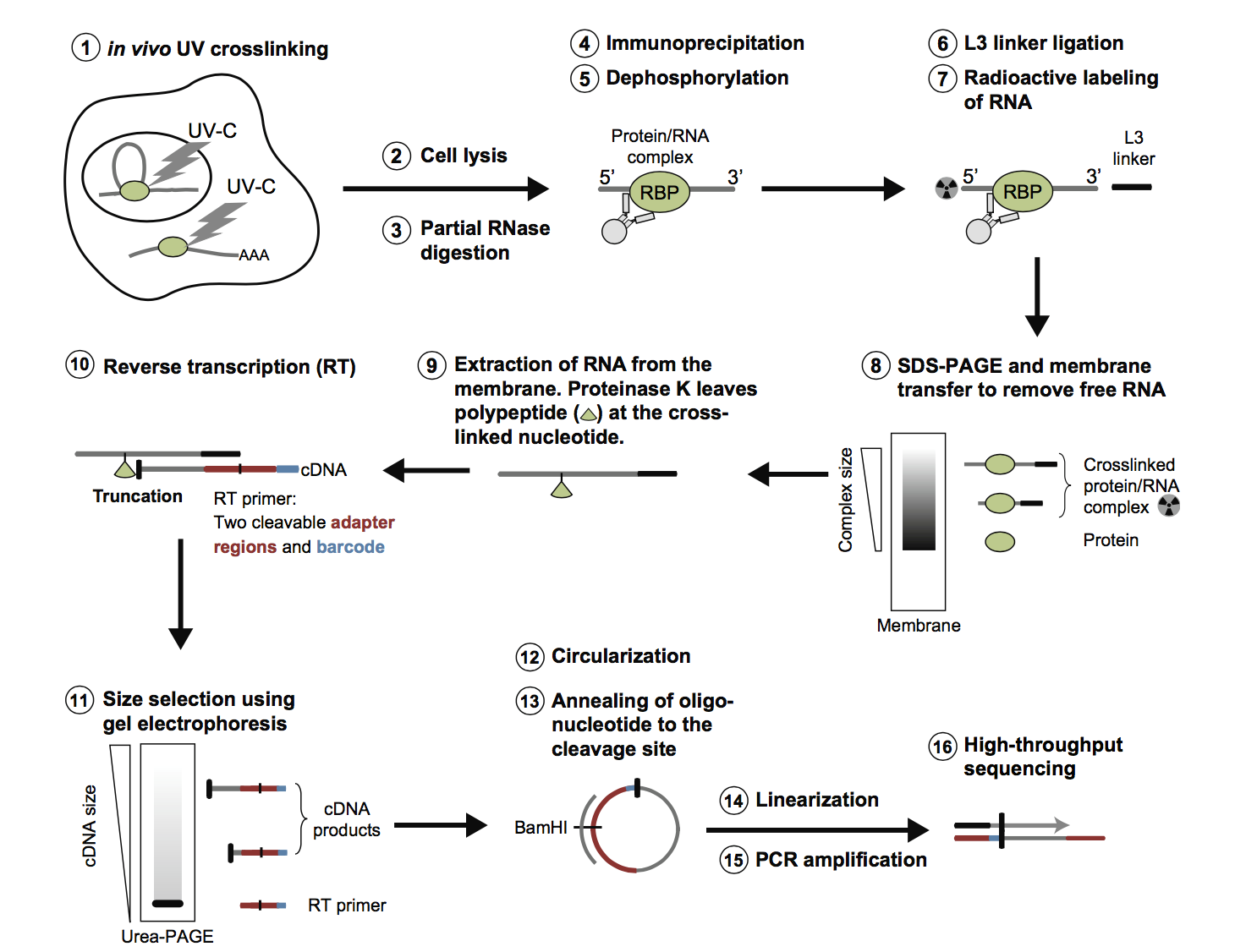
![PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e504d75c2c48b19613a796fb54083b71e3781289/2-Figure1-1.png)




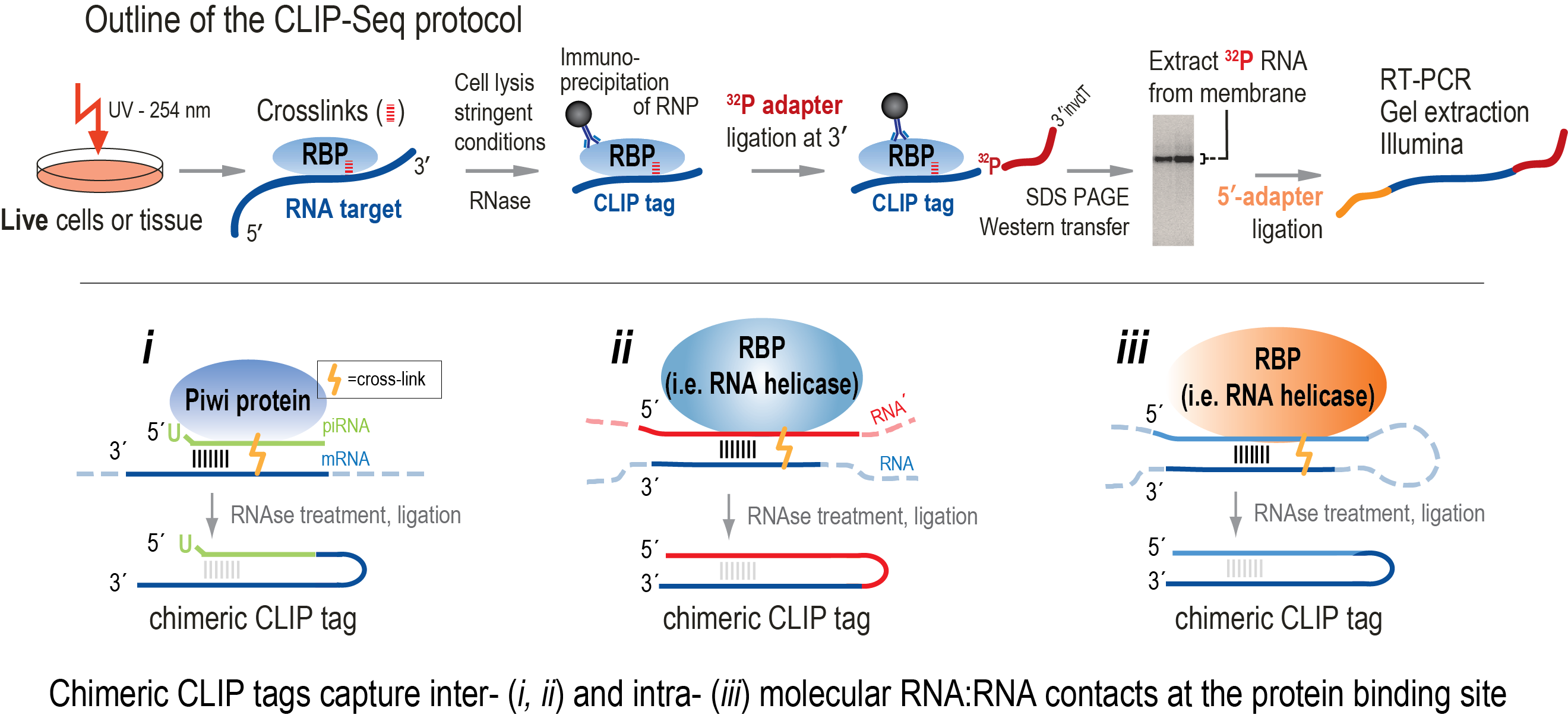
![PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e504d75c2c48b19613a796fb54083b71e3781289/3-Figure2-1.png)