3 Summary model of CMG loading and activation during DNA replication.... | Download Scientific Diagram

Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase | PNAS

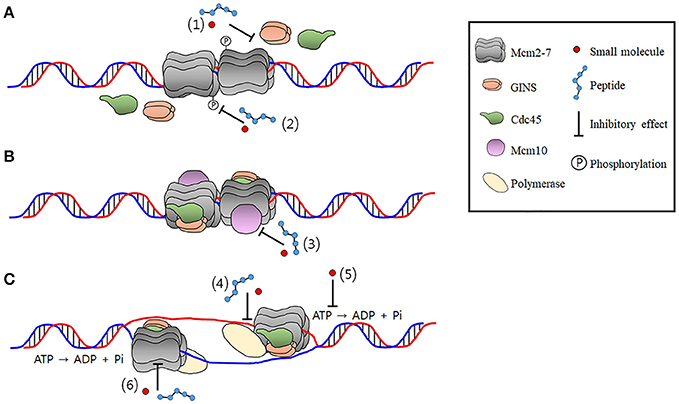
Myc and the Replicative CMG Helicase: The Creation and Destruction of Cancer - Reed - 2020 - BioEssays - Wiley Online Library

Structure of a human replisome shows the organisation and interactions of a DNA replication machine | The EMBO Journal

Mechanisms in helicase activation and analysis of the replication fork activity and architecture – Speck Lab / DNA Replication Group – Christian Speck
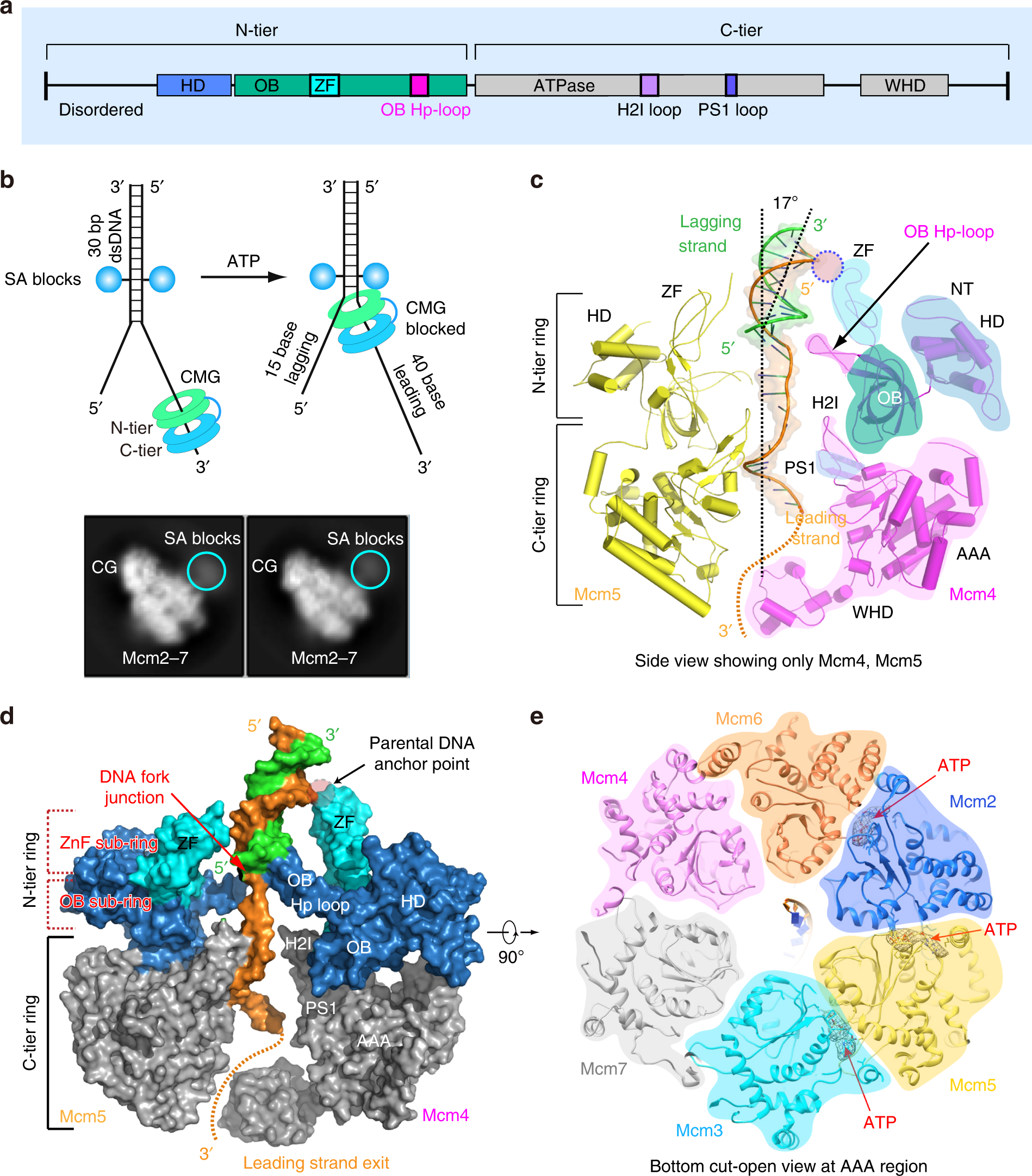
Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate | Nature Communications

Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication | Science
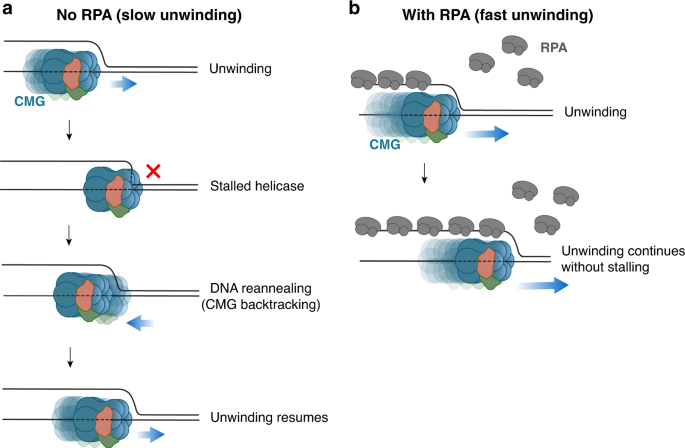
Duplex DNA engagement and RPA oppositely regulate the DNA-unwinding rate of CMG helicase | Nature Communications

Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. - Abstract - Europe PMC

A model for hGINS at the replication fork. The hGINS tetramer is part... | Download Scientific Diagram

Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation | PNAS

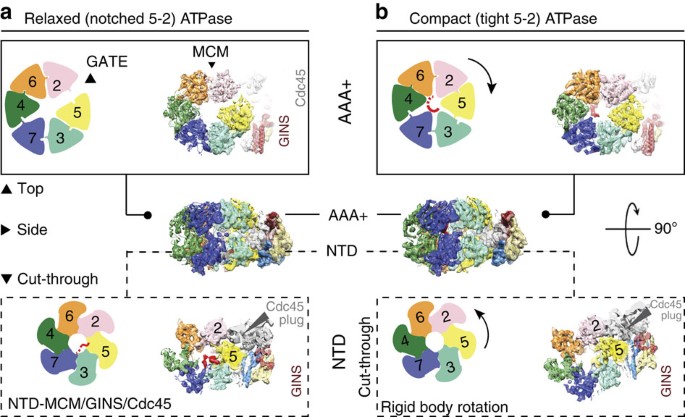
Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase - ScienceDirect
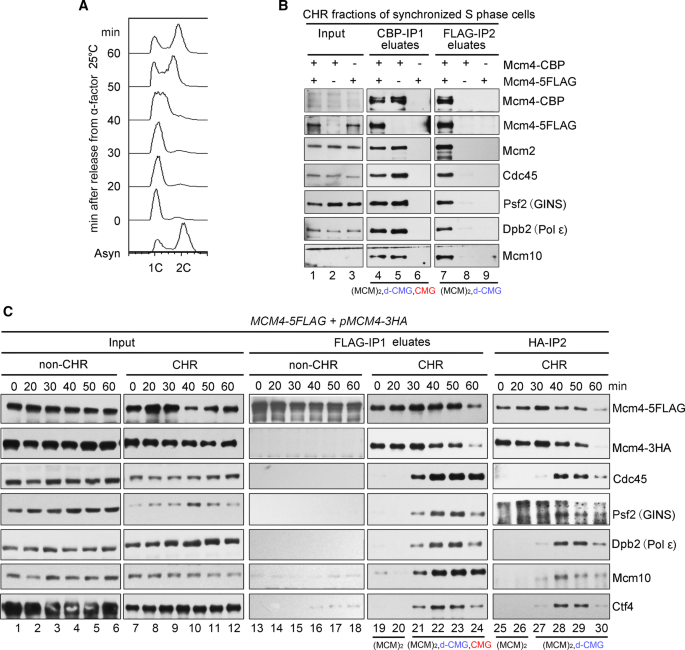
Characterization of the dimeric CMG/pre-initiation complex and its transition into DNA replication forks | SpringerLink
Structural components of the replisome. The CMG replicative helicase... | Download Scientific Diagram

Identification of the CMG complex in T. brucei. A, interactions among... | Download Scientific Diagram

CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication | PNAS