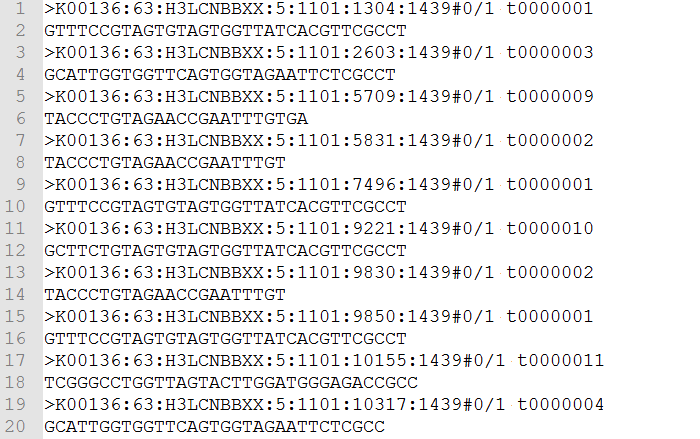
GitHub - robertaboukhalil/fastq.bio: An interactive web tool for quality control of DNA sequencing data

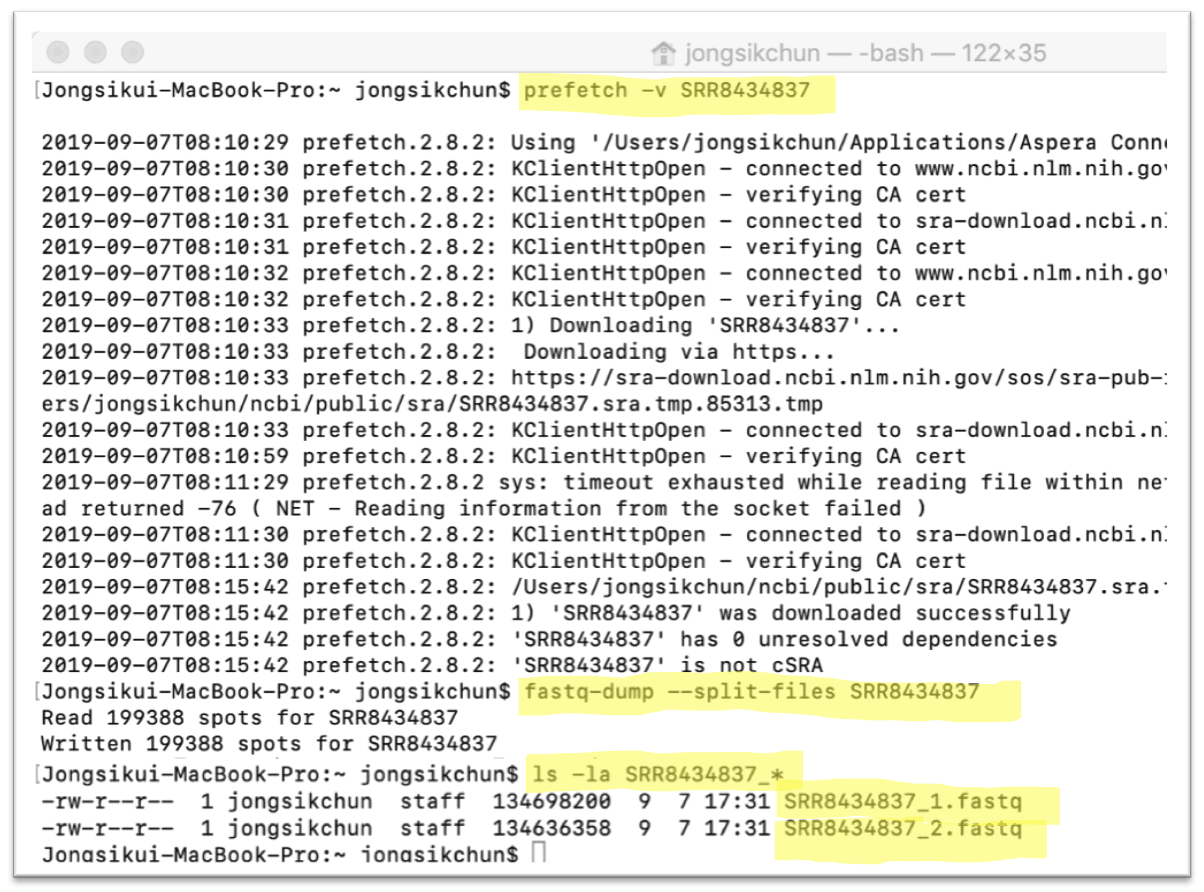
How long does it take to create single-end and pair-end data (fastq-dump) from SRAs? - usegalaxy.org support - Galaxy Community Help
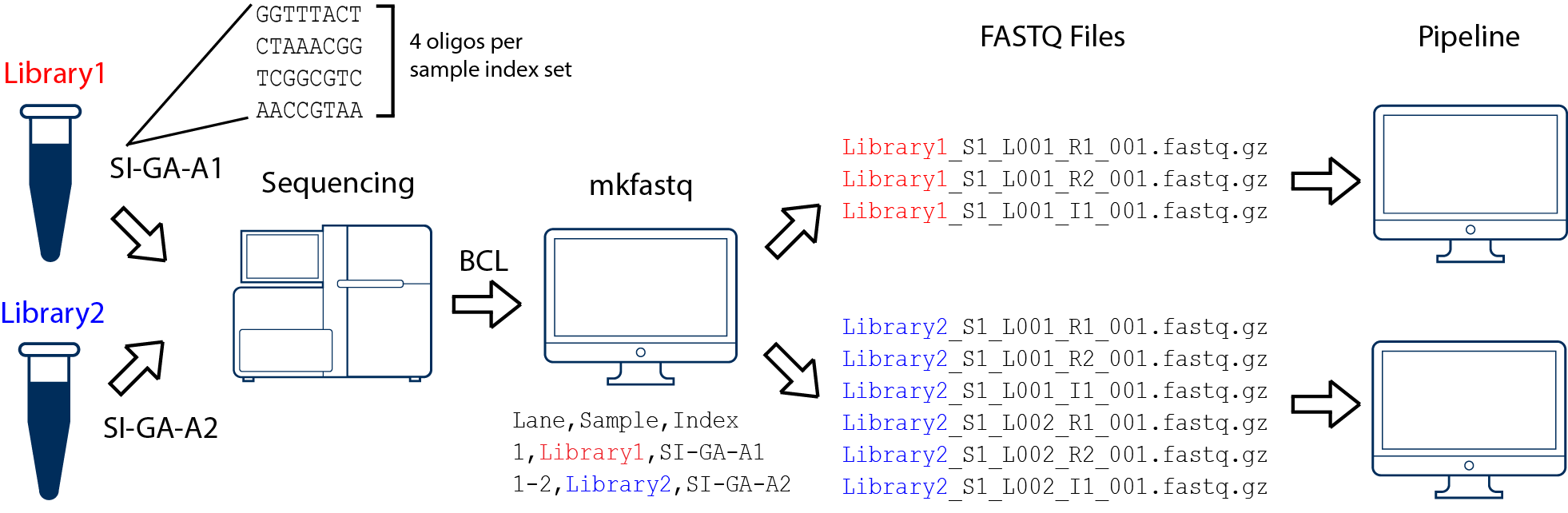
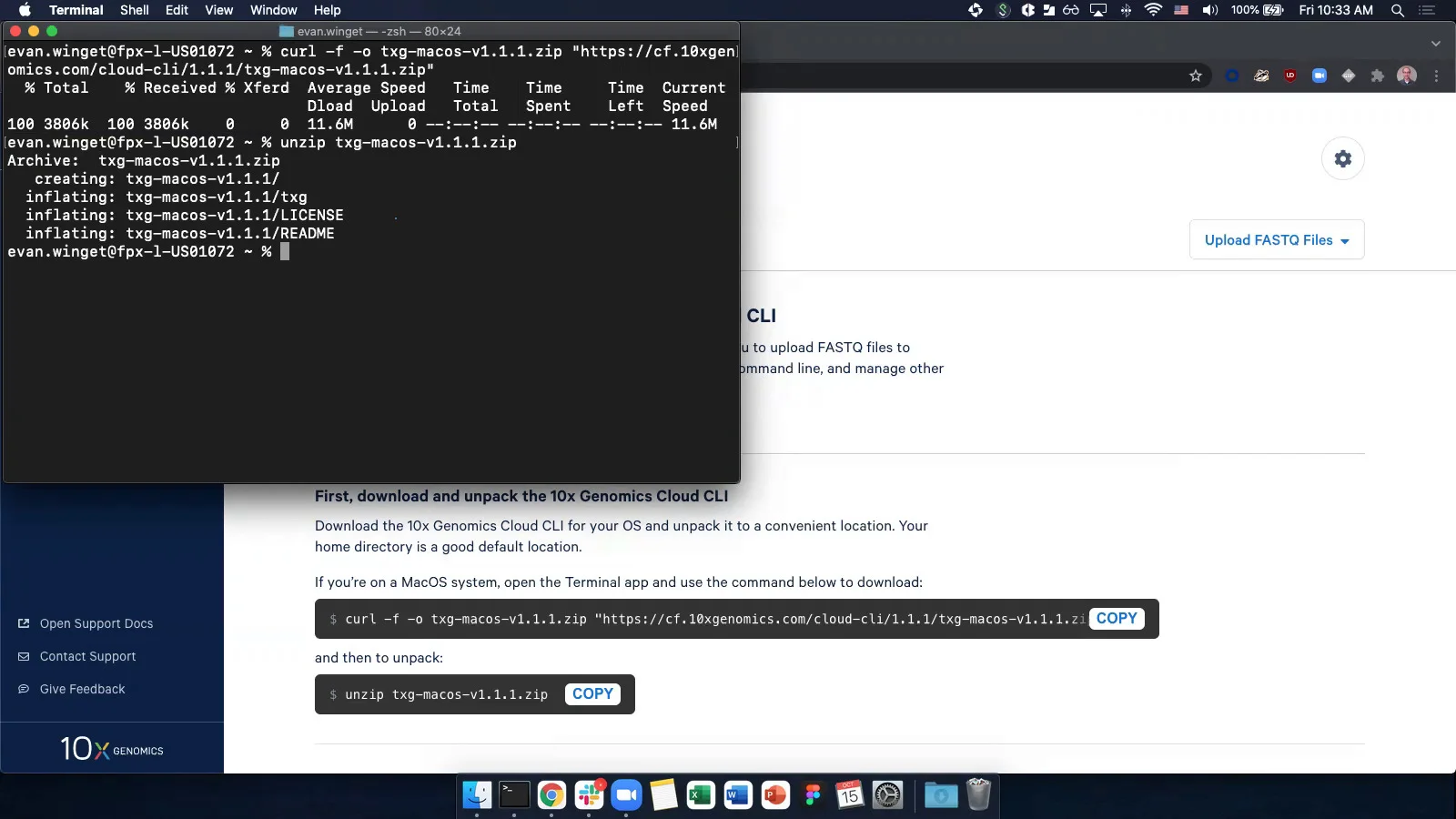
Generating FASTQs with cellranger mkfastq -Software -Single Cell Gene Expression -Official 10x Genomics Support
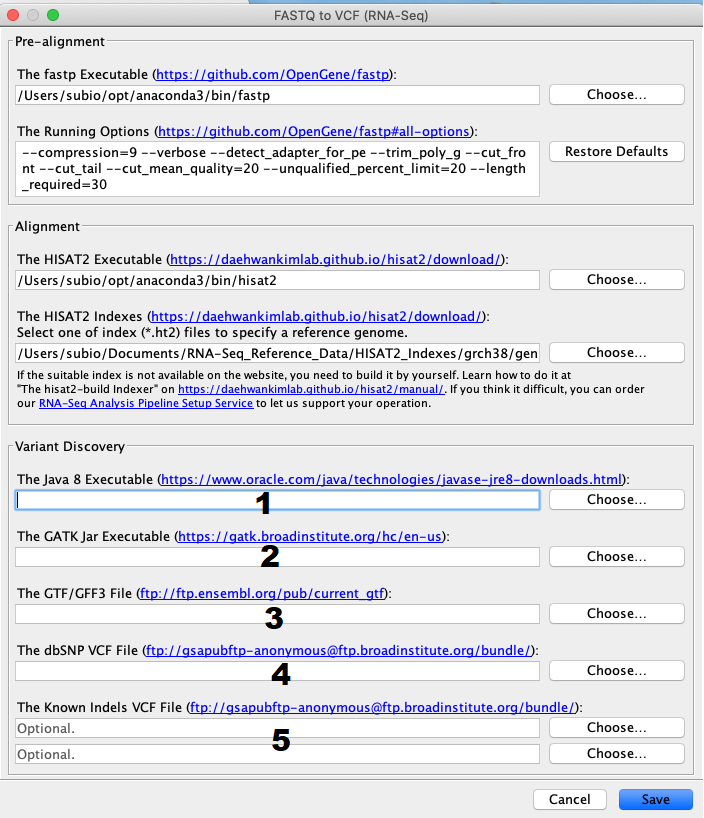
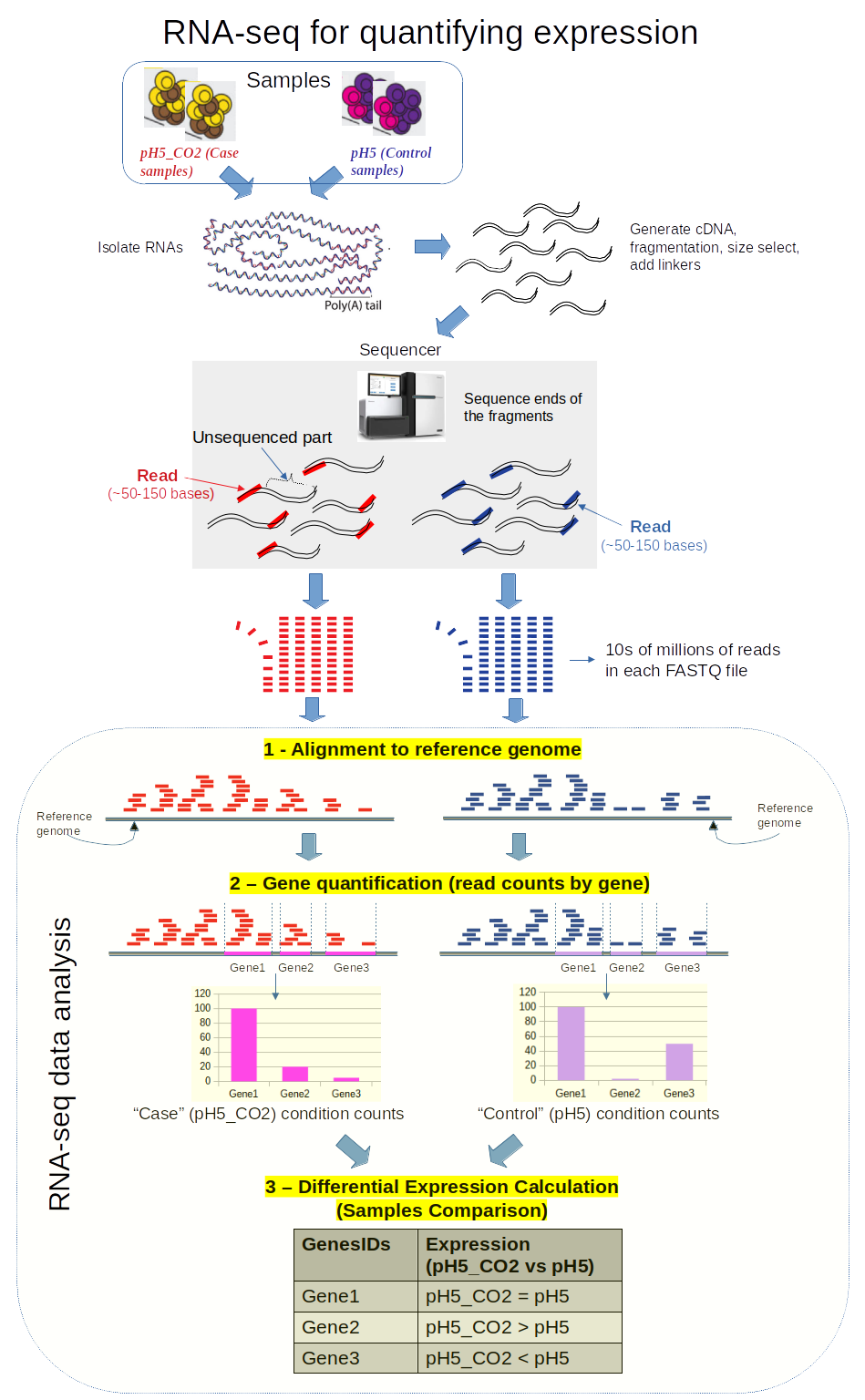

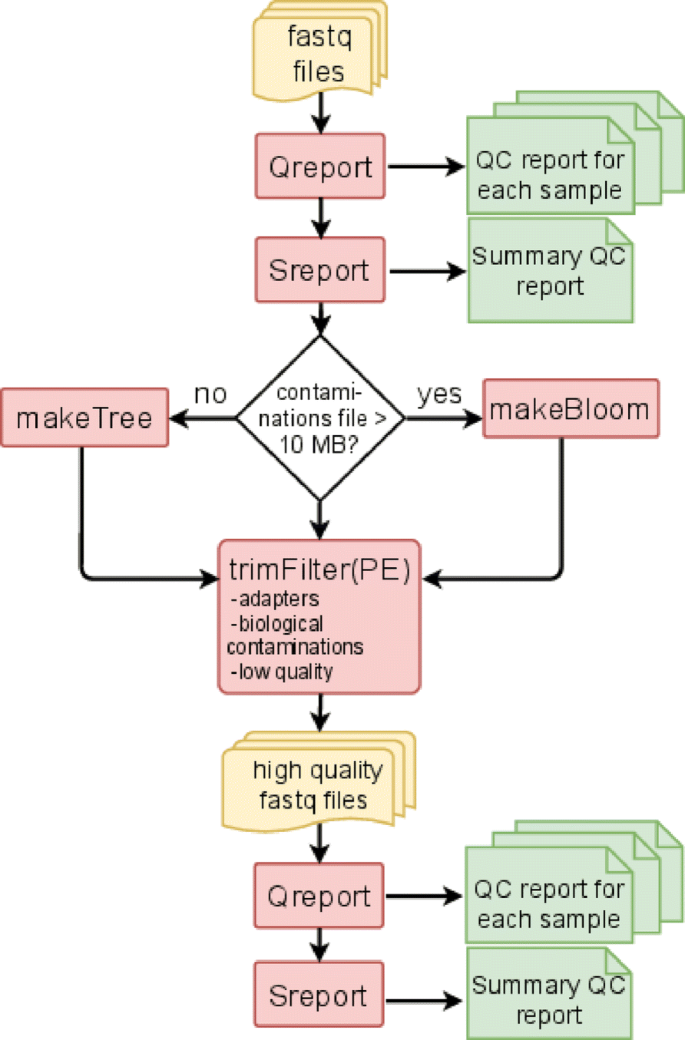
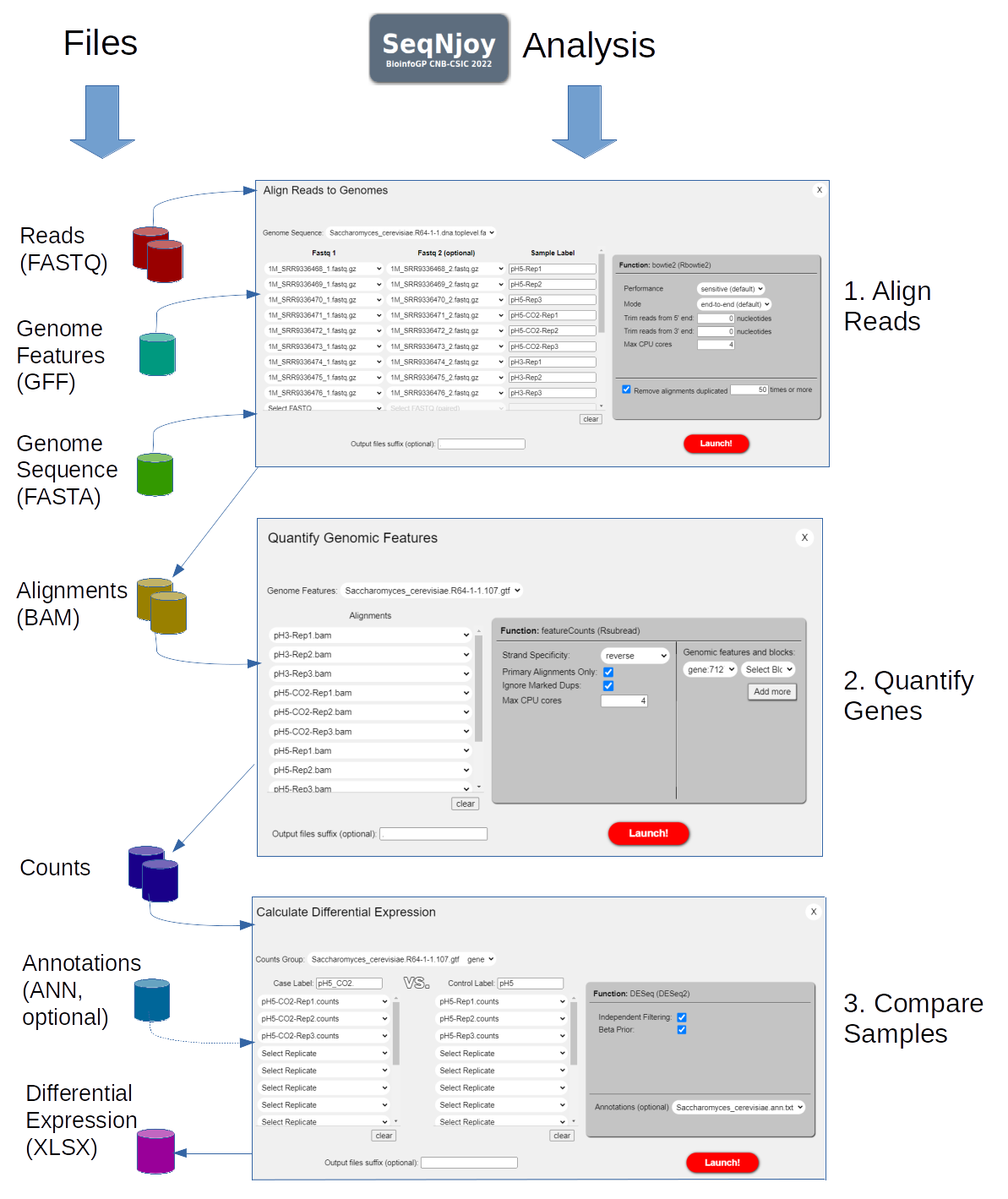







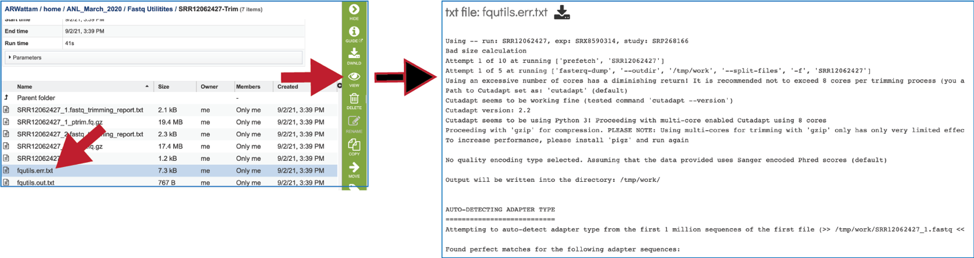
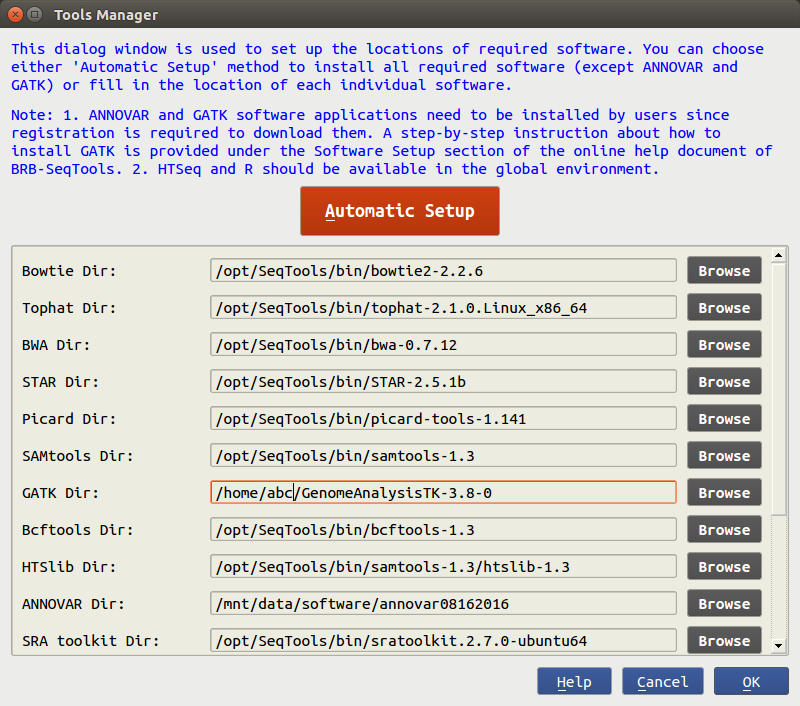
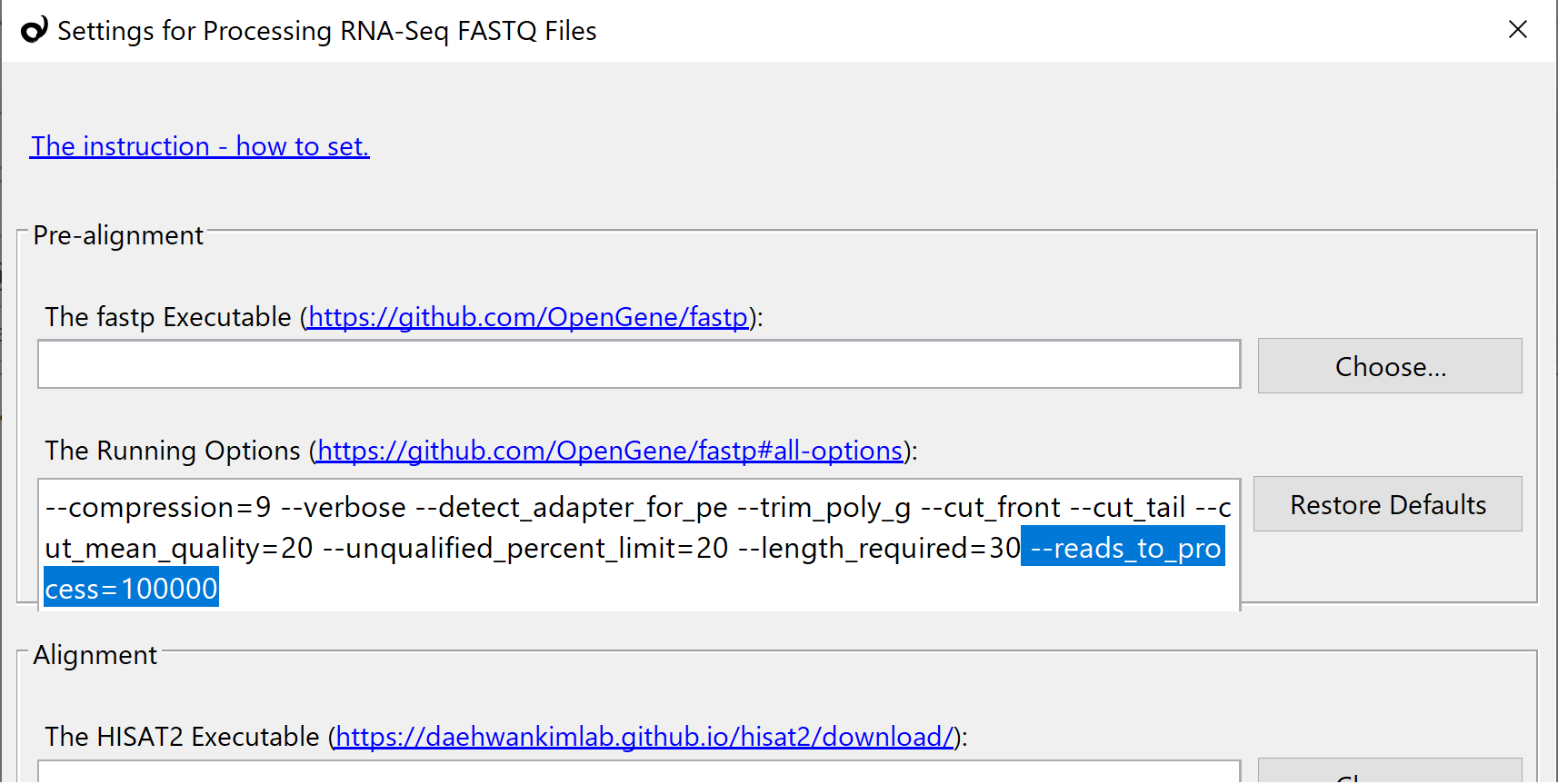
![2FAST2Q: a general-purpose sequence search and counting program for FASTQ files [PeerJ] 2FAST2Q: a general-purpose sequence search and counting program for FASTQ files [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/14041/1/fig-2-2x.jpg)