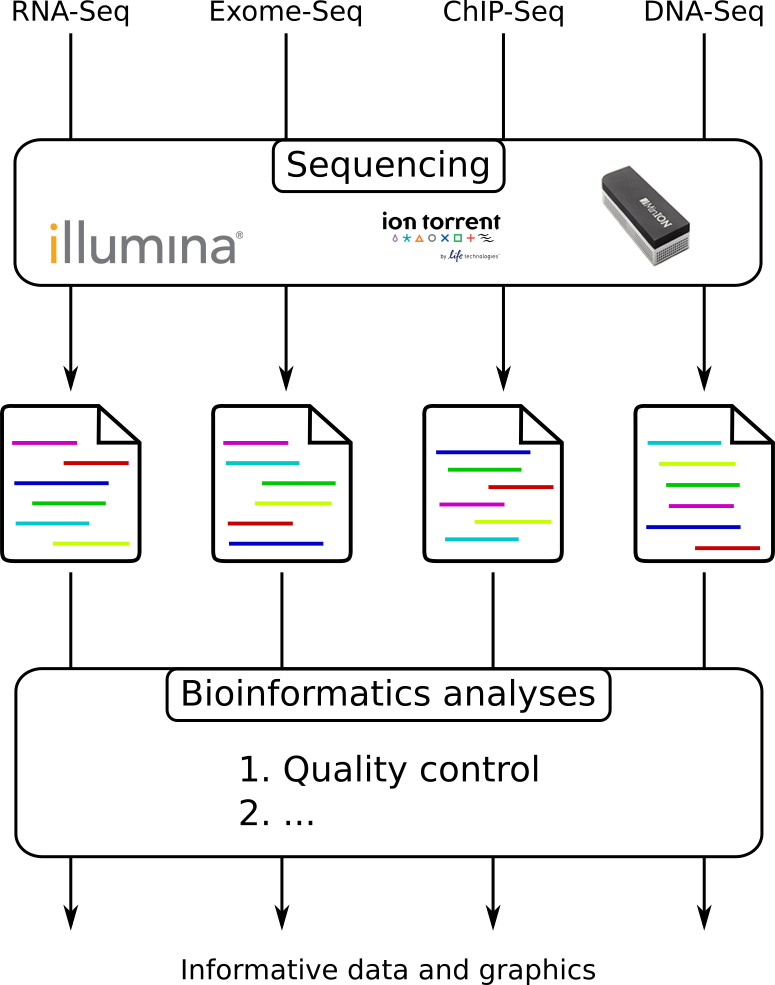
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics
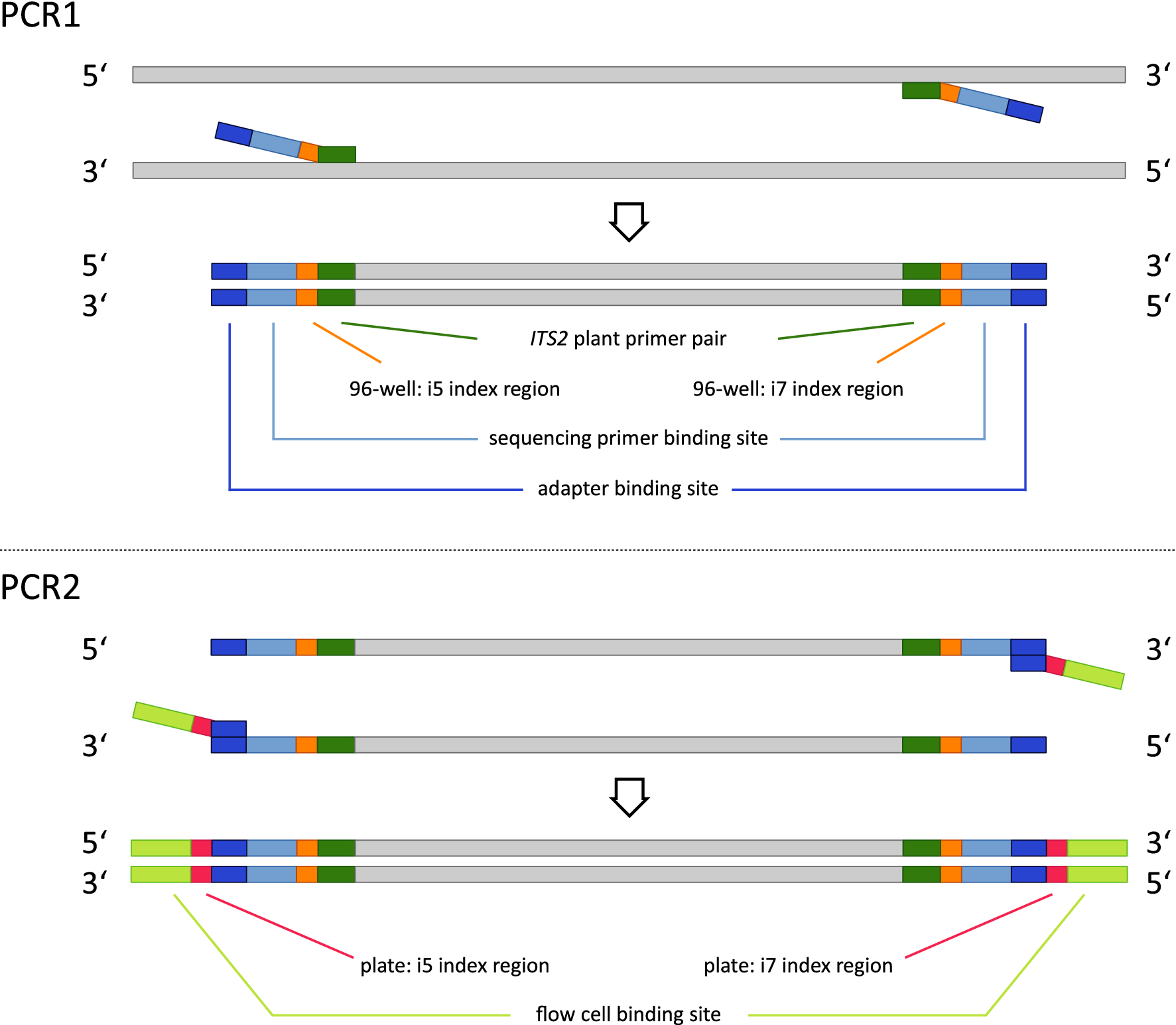
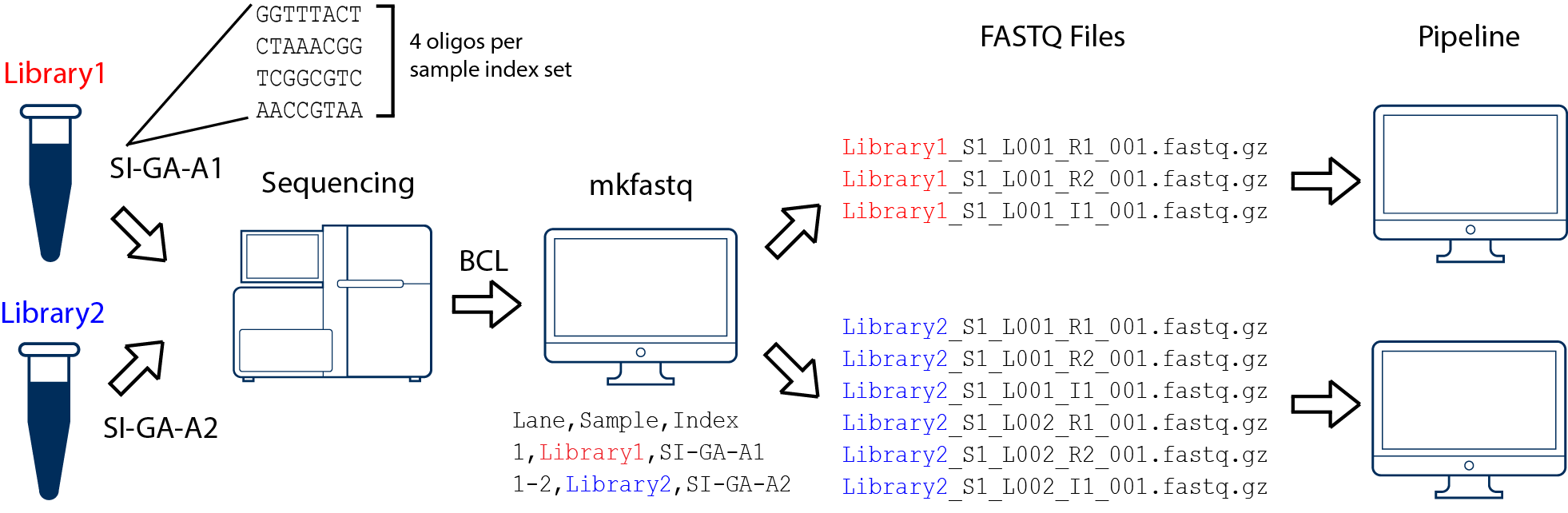
Handling of targeted amplicon sequencing data focusing on index hopping and demultiplexing using a nested metabarcoding approach in ecology | Scientific Reports
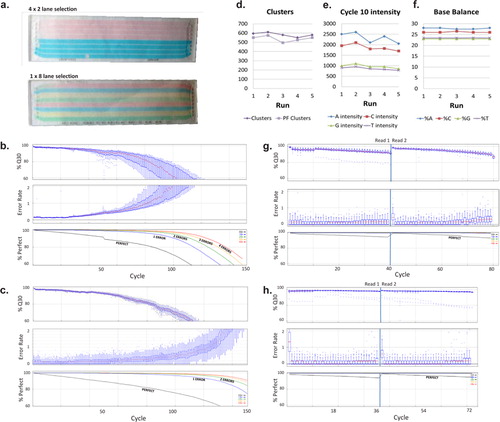
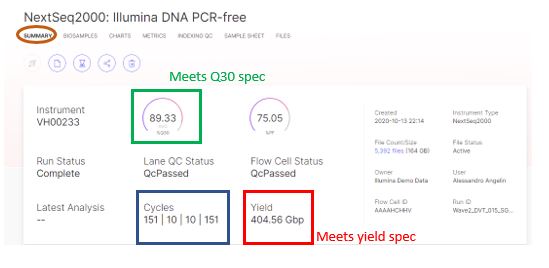
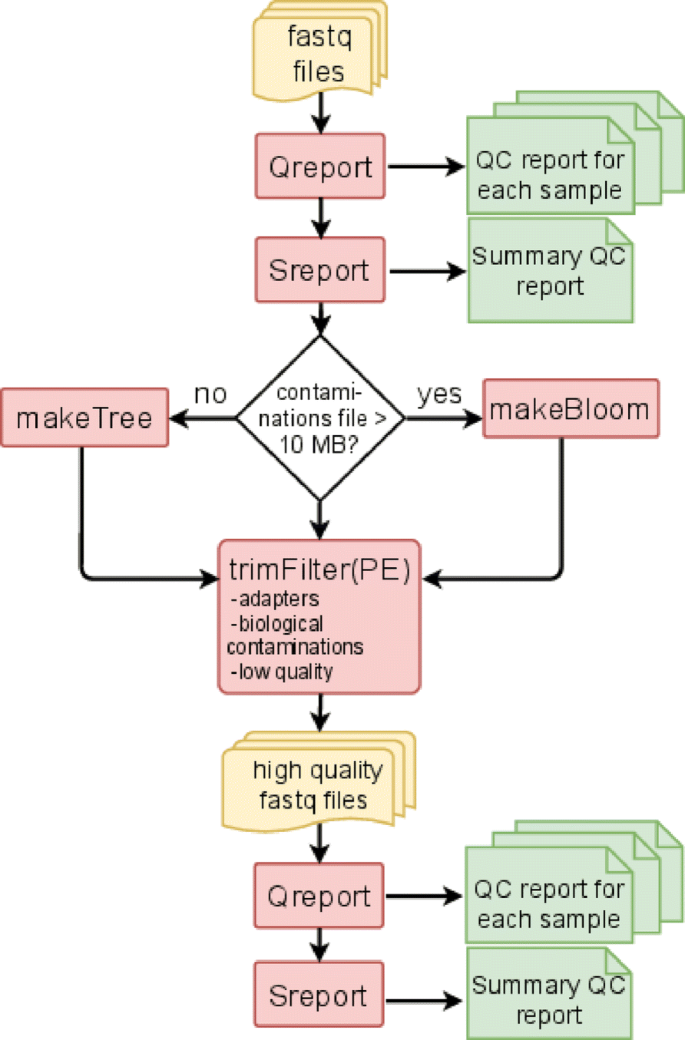
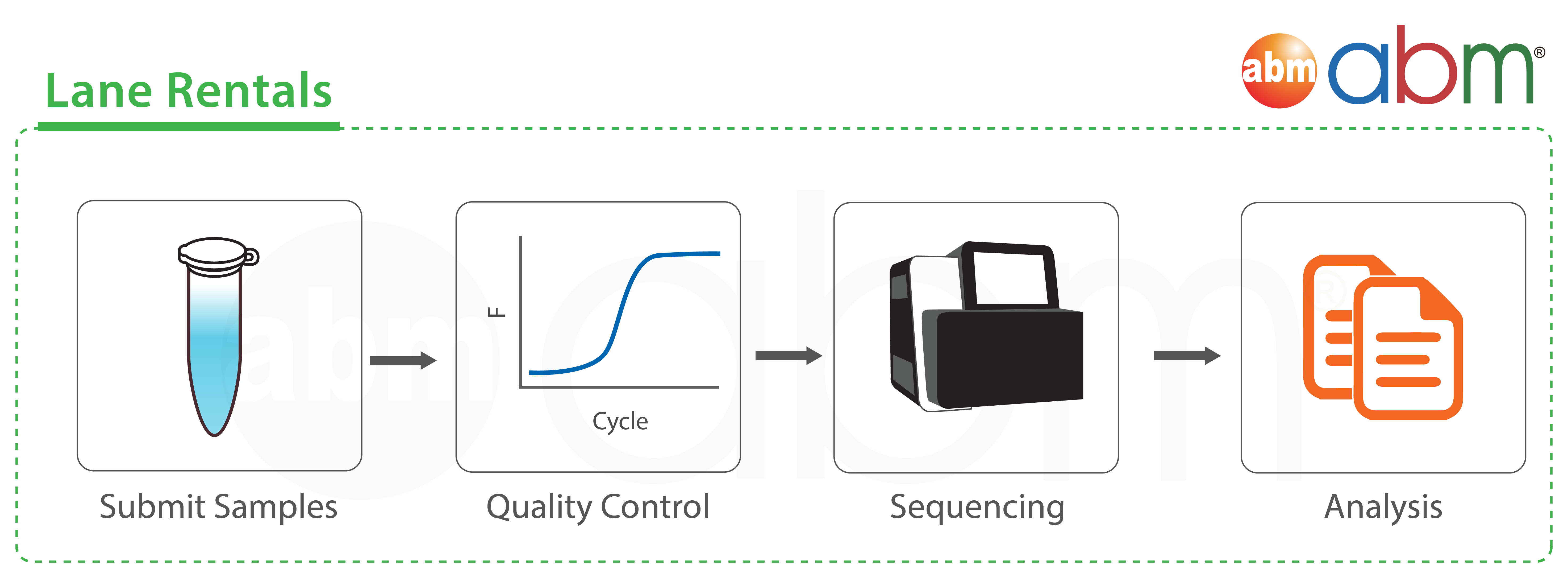
Our Top 5 Quality Control (QC) Metrics Every NGS User Should Know NGS QC metrics: 1. Sample Quality Control: 2. Library Qual

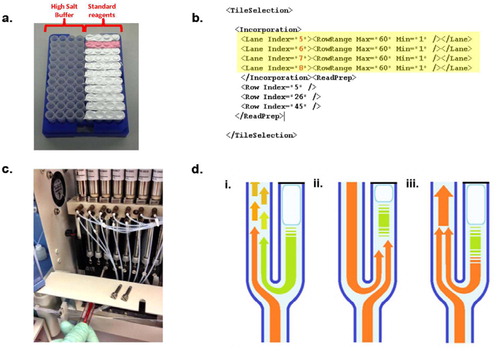
An open source toolkit for repurposing Illumina sequencing systems as versatile fluidics and imaging platforms | Scientific Reports

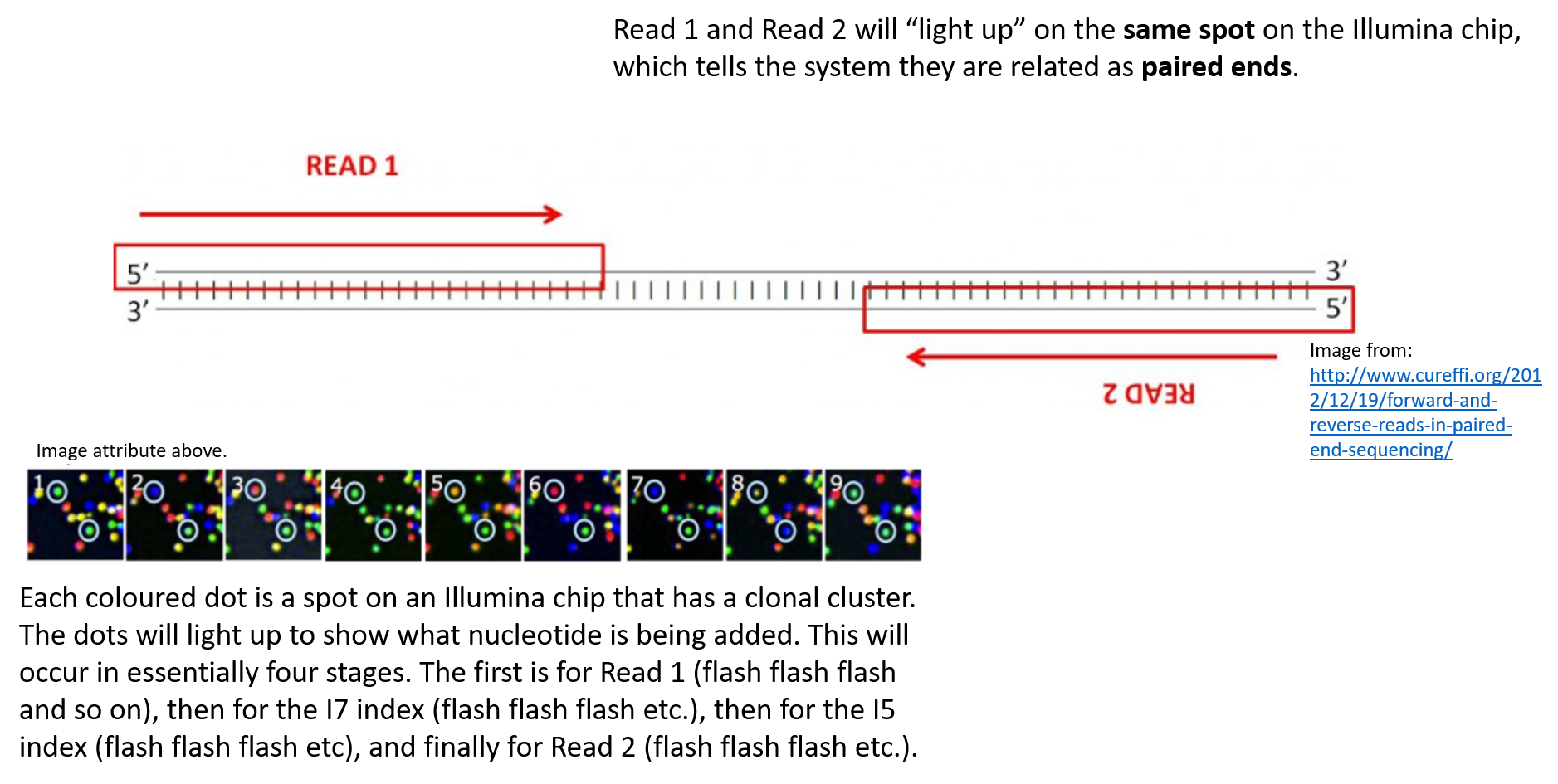
Comparison of Sanger sequencing (left) and Illumina next-generation... | Download Scientific Diagram