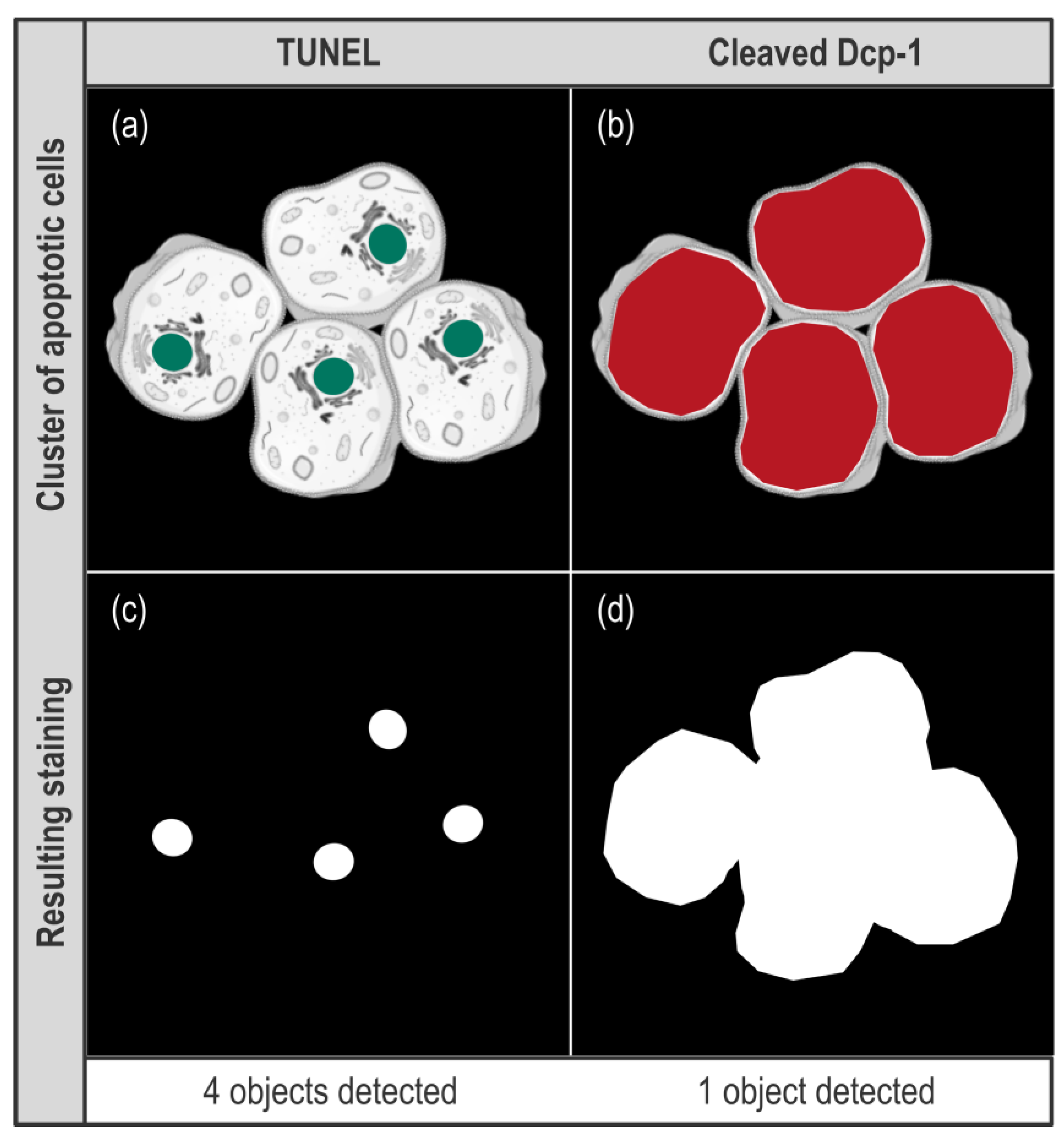
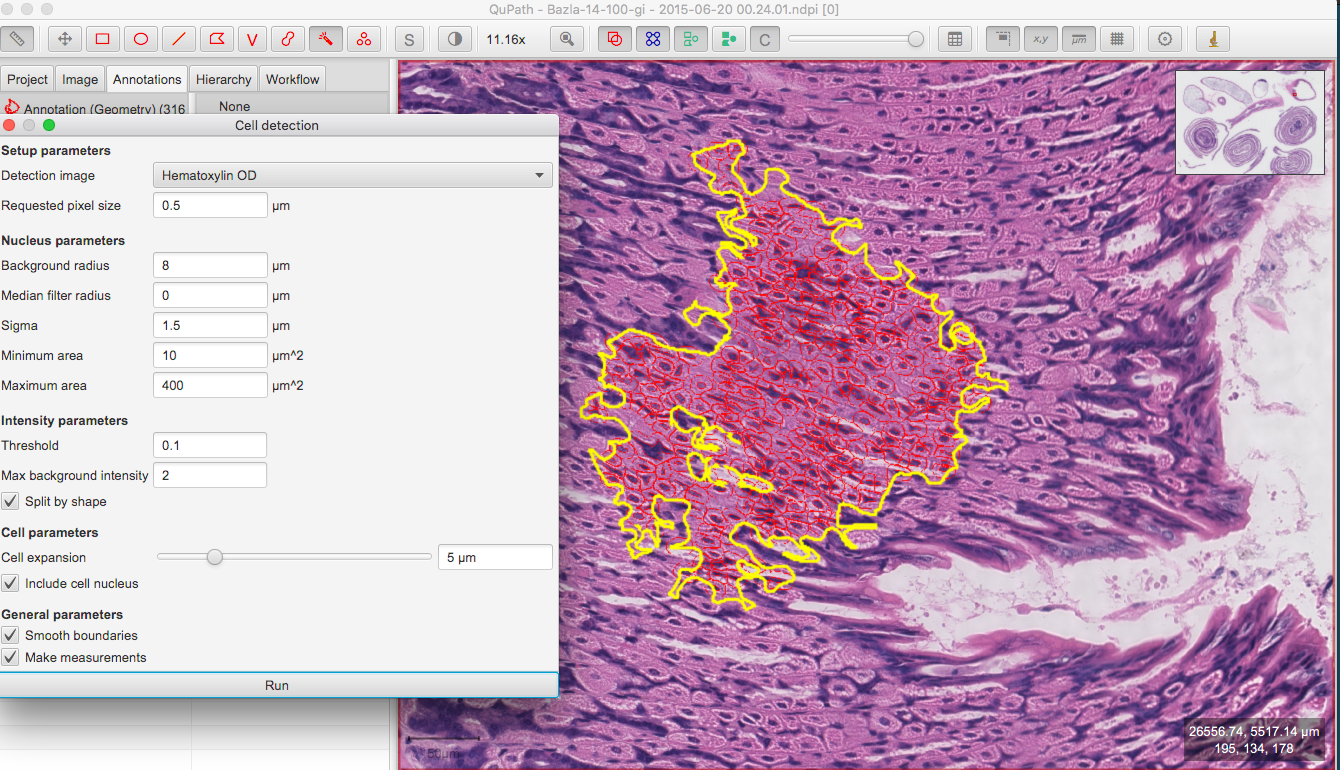
PDF) Whole-Slide Image Analysis of Human Pancreas Samples to Elucidate the Immunopathogenesis of Type 1 Diabetes Using the QuPath Software
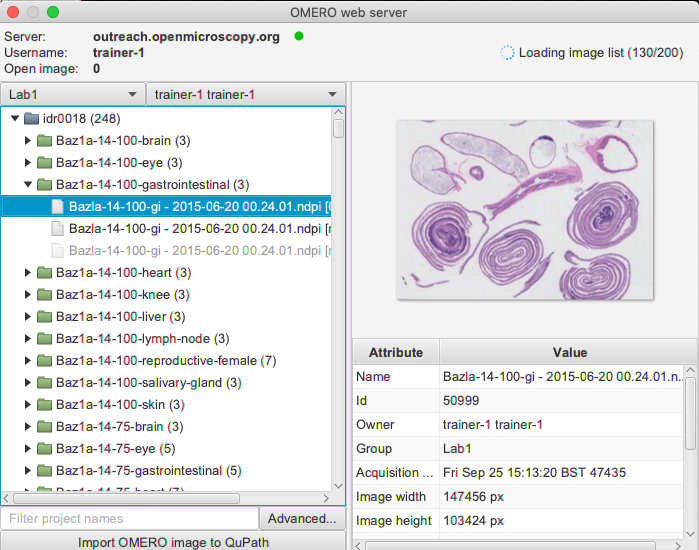
tutorials/monailabel_pathology_nuclei_segmentation_QuPath.ipynb at main · Project-MONAI/tutorials · GitHub
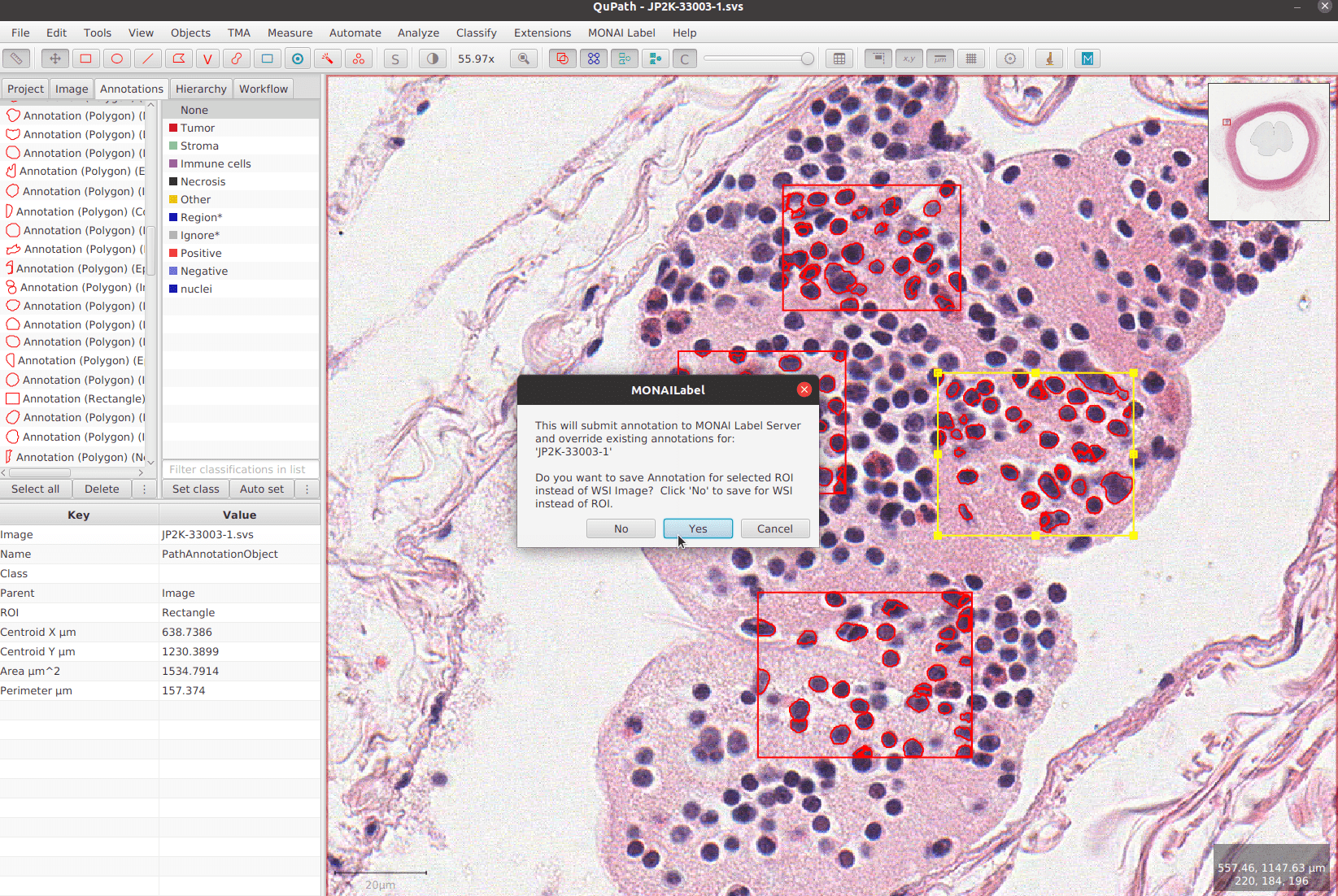
tutorials/monailabel_pathology_nuclei_segmentation_QuPath.ipynb at main · Project-MONAI/tutorials · GitHub

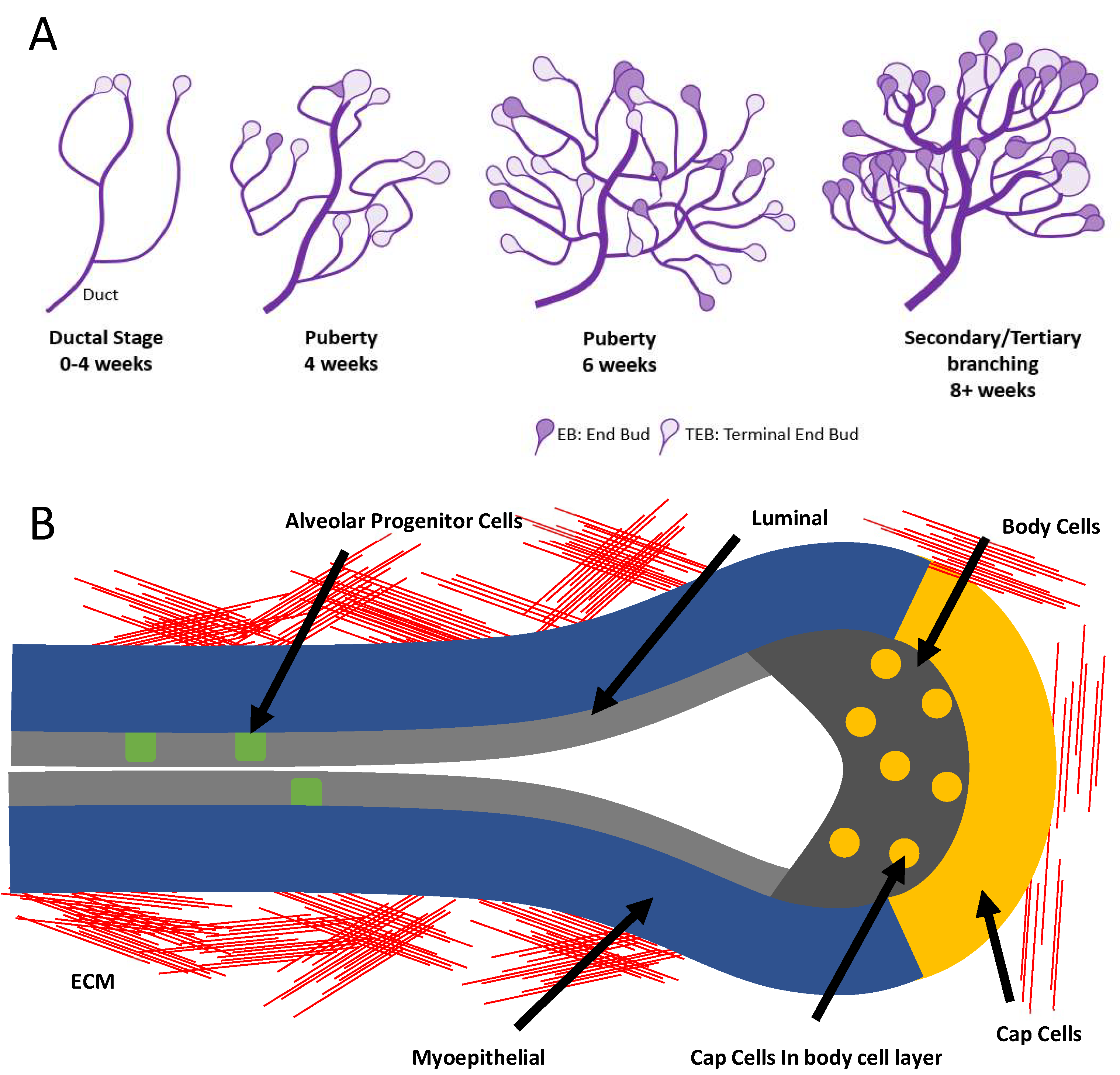
Olfactory bulb astrocytes mediate sensory circuit processing through Sox9 in the mouse brain | Nature Communications
PDF) Whole-Slide Image Analysis of Human Pancreas Samples to Elucidate the Immunopathogenesis of Type 1 Diabetes Using the QuPath Software

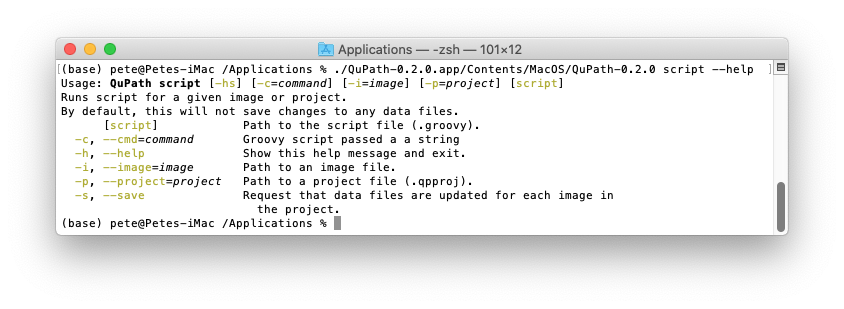
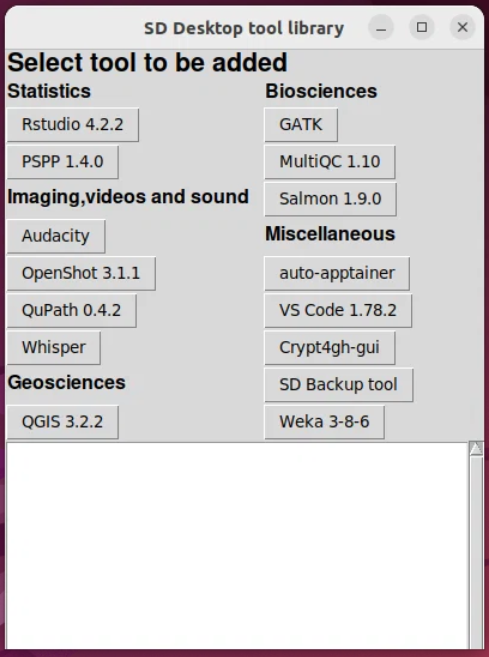
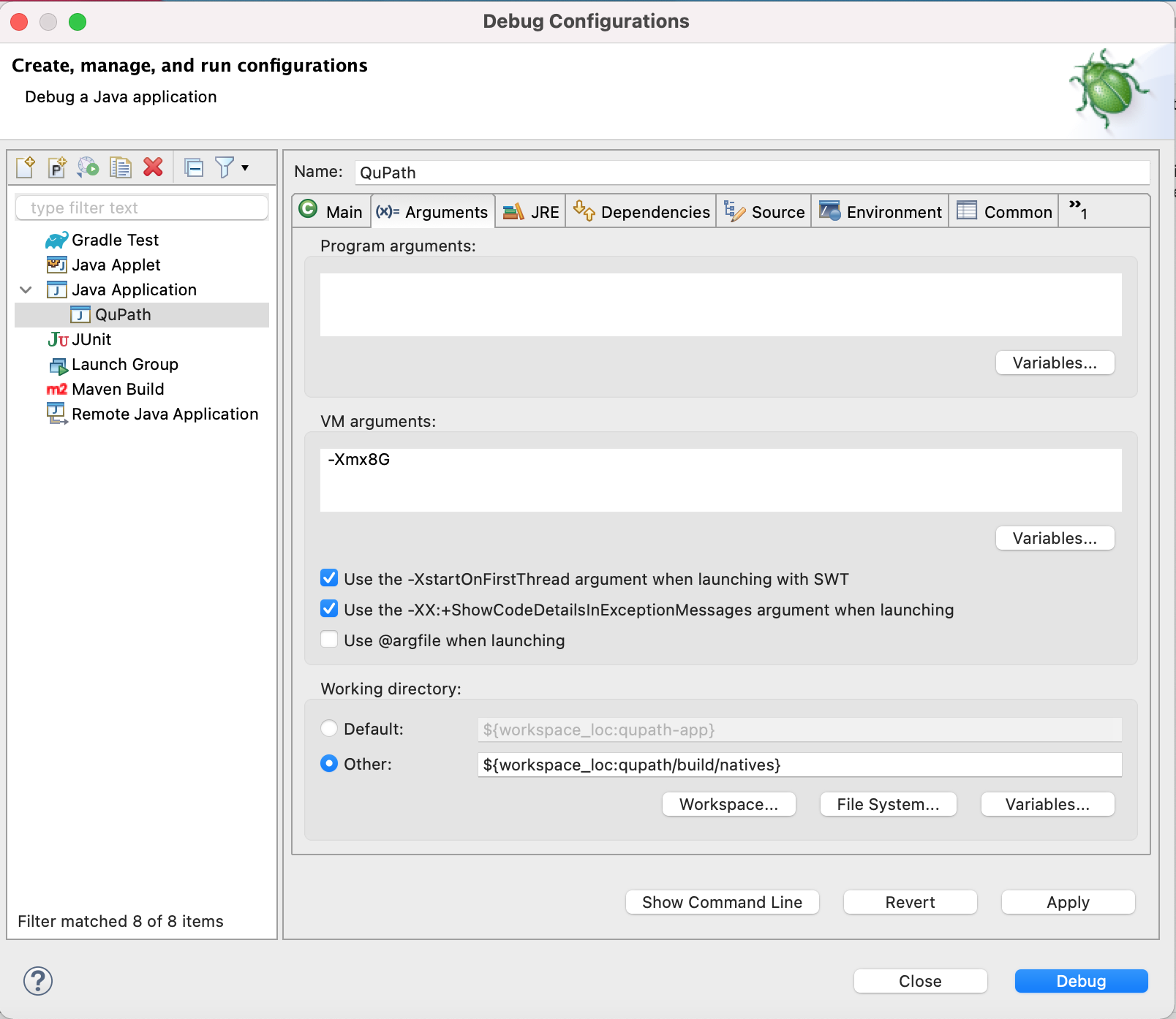
I would like to add a section in the documentation on developing QuPath with Visual Studio Code. Any help, suggestions, testing appreciated! - Development - Image.sc Forum

I would like to add a section in the documentation on developing QuPath with Visual Studio Code. Any help, suggestions, testing appreciated! - Development - Image.sc Forum